diff --git a/.github/workflows/test.yml b/.github/workflows/test.yml
index 17dcc3ee..db3e1fbc 100644
--- a/.github/workflows/test.yml
+++ b/.github/workflows/test.yml
@@ -11,7 +11,7 @@ jobs:
strategy:
matrix:
python-version: [3.7, 3.8, 3.9]
- os: [ubuntu-latest, windows-latest]
+ os: [ubuntu-latest]
steps:
- uses: actions/checkout@v2
@@ -21,8 +21,13 @@ jobs:
with:
python-version: ${{ matrix.python-version }}
- - name: Install package
- run: pip install .
+ - name: Install dev-package
+ run: |
+ sudo apt-get install qemu tree
+ python -m pip install --upgrade pip
+ pip install -v -e .
+ qemu-x86_64 -R 20M python time_match_strings.py
+
- name: Run tests
run: python -m unittest
diff --git a/CHANGELOG.md b/CHANGELOG.md
index d1cb63ff..7b77f8bd 100644
--- a/CHANGELOG.md
+++ b/CHANGELOG.md
@@ -7,6 +7,33 @@ and this project adheres to [Semantic Versioning](https://semver.org/spec/v2.0.0
## [Unreleased]
+## [0.6.0?] - 2021-09-21
+
+### Added
+
+* matrix-blocking/splitting as a performance-enhancer (see [README.md](https://github.com/ParticularMiner/string_grouper/tree/block#performance) for details)
+* new keyword arguments `force_symmetries` and `n_blocks` (see [README.md](https://github.com/ParticularMiner/string_grouper/tree/block#kwargs) for details)
+* new dependency on packages `topn` and `sparse_dot_topn_for_blocks` to help with the matrix-blocking
+* capability to reuse a previously initialized StringGrouper (that is, the corpus can now persist across high-level function calls like `match_strings()`. See [README.md](https://github.com/ParticularMiner/string_grouper/tree/block#corpus) for details.)
+
+
+## [0.5.0] - 2021-06-11
+
+### Added
+
+* Added new keyword argument **`tfidf_matrix_dtype`** (the datatype for the tf-idf values of the matrix components). Allowed values are `numpy.float32` and `numpy.float64` (used by the required external package `sparse_dot_topn` version 0.3.1). Default is `numpy.float32`. (Note: `numpy.float32` often leads to faster processing and a smaller memory footprint albeit less numerical precision than `numpy.float64`.)
+
+### Changed
+
+* Changed dependency on `sparse_dot_topn` from version 0.2.9 to 0.3.1
+* Changed the default datatype for cosine similarities from numpy.float64 to numpy.float32 to boost computational performance at the expense of numerical precision.
+* Changed the default value of the keyword argument `max_n_matches` from 20 to the number of strings in `duplicates` (or `master`, if `duplicates` is not given).
+* Changed warning issued when the condition \[`include_zeroes=True` and `min_similarity` ≤ 0 and `max_n_matches` is not sufficiently high to capture all nonzero-similarity-matches\] is met to an exception.
+
+### Removed
+
+* Removed the keyword argument `suppress_warning`
+
## [0.4.0] - 2021-04-11
### Added
diff --git a/README.md b/README.md
index 13f22127..270b4e26 100644
--- a/README.md
+++ b/README.md
@@ -13,7 +13,7 @@
The image displayed above is a visualization of the graph-structure of one of the groups of strings found by `string_grouper`. Each circle (node) represents a string, and each connecting arc (edge) represents a match between a pair of strings with a similarity score above a given threshold score (here `0.8`).
-The ***centroid*** of the group, as determined by `string_grouper` (see [tutorials/group_representatives.md](tutorials/group_representatives.md) for an explanation), is the largest node, also with the most edges originating from it. A thick line in the image denotes a strong similarity between the nodes at its ends, while a faint thin line denotes weak similarity.
+The ***centroid*** of the group, as determined by `string_grouper` (see [tutorials/group_representatives.md](https://github.com/Bergvca/string_grouper/blob/master/tutorials/group_representatives.md) for an explanation), is the largest node, also with the most edges originating from it. A thick line in the image denotes a strong similarity between the nodes at its ends, while a faint thin line denotes weak similarity.
The power of `string_grouper` is discernible from this image: in large datasets, `string_grouper` is often able to resolve indirect associations between strings even when, say, due to memory-resource-limitations, direct matches between those strings cannot be computed using conventional methods with a lower threshold similarity score.
@@ -70,6 +70,18 @@ In the rest of this document the names, `Series` and `DataFrame`, refer to the f
|**`string_series_1(_2)`** | A `Series` of strings each of which is to be compared with its corresponding string in `string_series_2(_1)`. |
|**`**kwargs`** | Keyword arguments (see [below](#kwargs)).|
+***New in version 0.6.0***: each of the high-level functions listed above also has a `StringGrouper` method counterpart of the same name and parameters. Calling such a method of any instance of `StringGrouper` will not rebuild the instance's underlying corpus to make string-comparisons but rather use it to perform the string-comparisons. The input Series to the method (`master`, `duplicates`, and so on) will thus be encoded, or transformed, into tf-idf matrices, using this corpus. For example:
+```python
+# Build a corpus using strings in the pandas Series master:
+sg = StringGrouper(master)
+# The following method-calls will compare strings first in
+# pandas Series new_master_1 and next in new_master_2
+# using the corpus already built above without rebuilding or
+# changing it in any way:
+matches1 = sg.match_strings(new_master_1)
+matches2 = sg.match_strings(new_master_2)
+```
+
#### Functions:
* #### `match_strings`
@@ -85,7 +97,7 @@ In the rest of this document the names, `Series` and `DataFrame`, refer to the f
2. `'similarity'` whose column has the similarity-scores as values, and
3. The name of `duplicates` (or `master` if `duplicates` is not given) and the name(s) of its index (or index-levels) prefixed by the string `'right_'`.
- Indexes (or their levels) only appear when the keyword argument `ignore_index=False` (the default). (See [tutorials/ignore_index_and_replace_na.md](tutorials/ignore_index_and_replace_na.md) for a demonstration.)
+ Indexes (or their levels) only appear when the keyword argument `ignore_index=False` (the default). (See [tutorials/ignore_index_and_replace_na.md](https://github.com/Bergvca/string_grouper/blob/master/tutorials/ignore_index_and_replace_na.md) for a demonstration.)
If either `master` or `duplicates` has no name, it assumes the name `'side'` which is then prefixed as described above. Similarly, if any of the indexes (or index-levels) has no name it assumes its `pandas` default name (`'index'`, `'level_0'`, and so on) and is then prefixed as described above.
@@ -101,7 +113,7 @@ In the rest of this document the names, `Series` and `DataFrame`, refer to the f
The name of the output `Series` is the same as that of `master` prefixed with the string `'most_similar_'`. If `master` has no name, it is assumed to have the name `'master'` before being prefixed.
- If `ignore_index=False` (the default), `match_most_similar` returns a `DataFrame` containing the same `Series` described above as one of its columns. So it inherits the same index and length as `duplicates`. The rest of its columns correspond to the index (or index-levels) of `master` and thus contain the index-labels of the most similar strings being output as values. If there are no similar strings in `master` for a given string in `duplicates` then the value(s) assigned to this index-column(s) for that string is `NaN` by default. However, if the keyword argument `replace_na=True`, then these `NaN` values are replaced with the index-label(s) of that string in `duplicates`. Note that such replacements can only occur if the indexes of `master` and `duplicates` have the same number of levels. (See [tutorials/ignore_index_and_replace_na.md](tutorials/ignore_index_and_replace_na.md#MMS) for a demonstration.)
+ If `ignore_index=False` (the default), `match_most_similar` returns a `DataFrame` containing the same `Series` described above as one of its columns. So it inherits the same index and length as `duplicates`. The rest of its columns correspond to the index (or index-levels) of `master` and thus contain the index-labels of the most similar strings being output as values. If there are no similar strings in `master` for a given string in `duplicates` then the value(s) assigned to this index-column(s) for that string is `NaN` by default. However, if the keyword argument `replace_na=True`, then these `NaN` values are replaced with the index-label(s) of that string in `duplicates`. Note that such replacements can only occur if the indexes of `master` and `duplicates` have the same number of levels. (See [tutorials/ignore_index_and_replace_na.md](https://github.com/Bergvca/string_grouper/blob/master/tutorials/ignore_index_and_replace_na.md#MMS) for a demonstration.)
Each column-name of the output `DataFrame` has the same name as its corresponding column, index, or index-level of `master` prefixed with the string `'most_similar_'`.
@@ -109,7 +121,7 @@ In the rest of this document the names, `Series` and `DataFrame`, refer to the f
* #### `group_similar_strings`
- Takes a single `Series` of strings (`strings_to_group`) and groups them by assigning to each string one string from `strings_to_group` chosen as the group-representative for each group of similar strings found. (See [tutorials/group_representatives.md](tutorials/group_representatives.md) for details on how the the group-representatives are chosen.)
+ Takes a single `Series` of strings (`strings_to_group`) and groups them by assigning to each string one string from `strings_to_group` chosen as the group-representative for each group of similar strings found. (See [tutorials/group_representatives.md](https://github.com/Bergvca/string_grouper/blob/master/tutorials/group_representatives.md) for details on how the the group-representatives are chosen.)
If `ignore_index=True`, the output is a `Series` (with the same name as `strings_to_group` prefixed by the string `'group_rep_'`) of the same length and index as `strings_to_group` containing the group-representative strings. If `strings_to_group` has no name then the name of the returned `Series` is `'group_rep'`.
@@ -134,17 +146,20 @@ All functions are built using a class **`StringGrouper`**. This class can be use
All keyword arguments not mentioned in the function definitions above are used to update the default settings. The following optional arguments can be used:
* **`ngram_size`**: The amount of characters in each n-gram. Default is `3`.
- * **`regex`**: The regex string used to clean-up the input string. Default is `"[,-./]|\s"`.
- * **`max_n_matches`**: The maximum number of matches allowed per string in `master`. Default is `20`.
+ * **`regex`**: The regex string used to clean-up the input string. Default is `r"[,-./]|\s"`.
+ * **`ignore_case`**: Determines whether or not letter case in strings should be ignored. Defaults to `True`.
+ * **`tfidf_matrix_dtype`**: The datatype for the tf-idf values of the matrix components. Allowed values are `numpy.float32` and `numpy.float64`. Default is `numpy.float32`. (Note: `numpy.float32` often leads to faster processing and a smaller memory footprint albeit less numerical precision than `numpy.float64`.)
+ * **`max_n_matches`**: The maximum number of matching strings in `master` allowed per string in `duplicates`. Default is the total number of strings in `master`.
* **`min_similarity`**: The minimum cosine similarity for two strings to be considered a match.
Defaults to `0.8`
* **`number_of_processes`**: The number of processes used by the cosine similarity calculation. Defaults to
`number of cores on a machine - 1.`
- * **`ignore_index`**: Determines whether indexes are ignored or not. If `False` (the default), index-columns will appear in the output, otherwise not. (See [tutorials/ignore_index_and_replace_na.md](tutorials/ignore_index_and_replace_na.md) for a demonstration.)
- * **`replace_na`**: For function `match_most_similar`, determines whether `NaN` values in index-columns are replaced or not by index-labels from `duplicates`. Defaults to `False`. (See [tutorials/ignore_index_and_replace_na.md](tutorials/ignore_index_and_replace_na.md) for a demonstration.)
- * **`include_zeroes`**: When `min_similarity` ≤ 0, determines whether zero-similarity matches appear in the output. Defaults to `True`. (See [tutorials/zero_similarity.md](tutorials/zero_similarity.md) for a demonstration.) **Warning:** Make sure the kwarg `max_n_matches` is sufficiently high to capture ***all*** nonzero-similarity-matches, otherwise some zero-similarity-matches returned will be false.
- * **`suppress_warning`**: when `min_similarity` ≤ 0 and `include_zeroes` is `True`, determines whether or not to suppress the message warning that `max_n_matches` may be too small. Defaults to `False`.
- * **`group_rep`**: For function `group_similar_strings`, determines how group-representatives are chosen. Allowed values are `'centroid'` (the default) and `'first'`. See [tutorials/group_representatives.md](tutorials/group_representatives.md) for an explanation.
+ * **`ignore_index`**: Determines whether indexes are ignored or not. If `False` (the default), index-columns will appear in the output, otherwise not. (See [tutorials/ignore_index_and_replace_na.md](https://github.com/Bergvca/string_grouper/blob/master/tutorials/ignore_index_and_replace_na.md) for a demonstration.)
+ * **`replace_na`**: For function `match_most_similar`, determines whether `NaN` values in index-columns are replaced or not by index-labels from `duplicates`. Defaults to `False`. (See [tutorials/ignore_index_and_replace_na.md](https://github.com/Bergvca/string_grouper/blob/master/tutorials/ignore_index_and_replace_na.md) for a demonstration.)
+ * **`include_zeroes`**: When `min_similarity` ≤ 0, determines whether zero-similarity matches appear in the output. Defaults to `True`. (See [tutorials/zero_similarity.md](https://github.com/Bergvca/string_grouper/blob/master/tutorials/zero_similarity.md).) **Note:** If `include_zeroes` is `True` and the kwarg `max_n_matches` is set then it must be sufficiently high to capture ***all*** nonzero-similarity-matches, otherwise an error is raised and `string_grouper` suggests an alternative value for `max_n_matches`. To allow `string_grouper` to automatically use the appropriate value for `max_n_matches` then do not set this kwarg at all.
+ * **`group_rep`**: For function `group_similar_strings`, determines how group-representatives are chosen. Allowed values are `'centroid'` (the default) and `'first'`. See [tutorials/group_representatives.md](https://github.com/Bergvca/string_grouper/blob/master/tutorials/group_representatives.md) for an explanation.
+ * **`force_symmetries`**: In cases where `duplicates` is `None`, specifies whether corrections should be made to the results to account for symmetry, thus compensating for those losses of numerical significance which violate the symmetries. Defaults to `True`.
+ * **`n_blocks`**: This parameter is a tuple of two `int`s provided to help boost performance, if possible, of processing large DataFrames (see [Subsection Performance](#perf)), by splitting the DataFrames into `n_blocks[0]` blocks for the left operand (of the underlying matrix multiplication) and into `n_blocks[1]` blocks for the right operand before performing the string-comparisons block-wise. Defaults to `None`, in which case automatic splitting occurs if an `OverflowError` would otherwise occur.
## Examples
@@ -306,7 +321,7 @@ Out of the four company names in `duplicates`, three companies are found in the
### Finding duplicates from a (database extract to) DataFrame where IDs for rows are supplied.
-A very common scenario is the case where duplicate records for an entity have been entered into a database. That is, there are two or more records where a name field has slightly different spelling. For example, "A.B. Corporation" and "AB Corporation". Using the optional 'ID' parameter in the `match_strings` function duplicates can be found easily. A [tutorial](tutorials/tutorial_1.md) that steps though the process with an example data set is available.
+A very common scenario is the case where duplicate records for an entity have been entered into a database. That is, there are two or more records where a name field has slightly different spelling. For example, "A.B. Corporation" and "AB Corporation". Using the optional 'ID' parameter in the `match_strings` function duplicates can be found easily. A [tutorial](https://github.com/Bergvca/string_grouper/blob/master/tutorials/tutorial_1.md) that steps though the process with an example data set is available.
### For a second data set, find only the most similar match
@@ -993,3 +1008,89 @@ companies[companies.deduplicated_name.str.contains('PRICEWATERHOUSECOOPERS LLP')
+
+# Performance
+
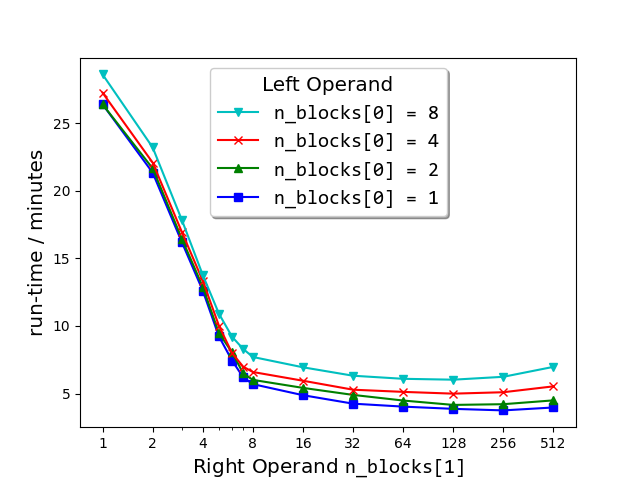
+### Semilogx plots of run-times of `match_strings()` vs the number of blocks (`n_blocks[1]`) into which the right matrix-operand of the dataset (663 000 strings from sec__edgar_company_info.csv) was split before performing the string comparison. As shown in the legend, each plot corresponds to the number `n_blocks[0]` of blocks into which the left matrix-operand was split.
+
+
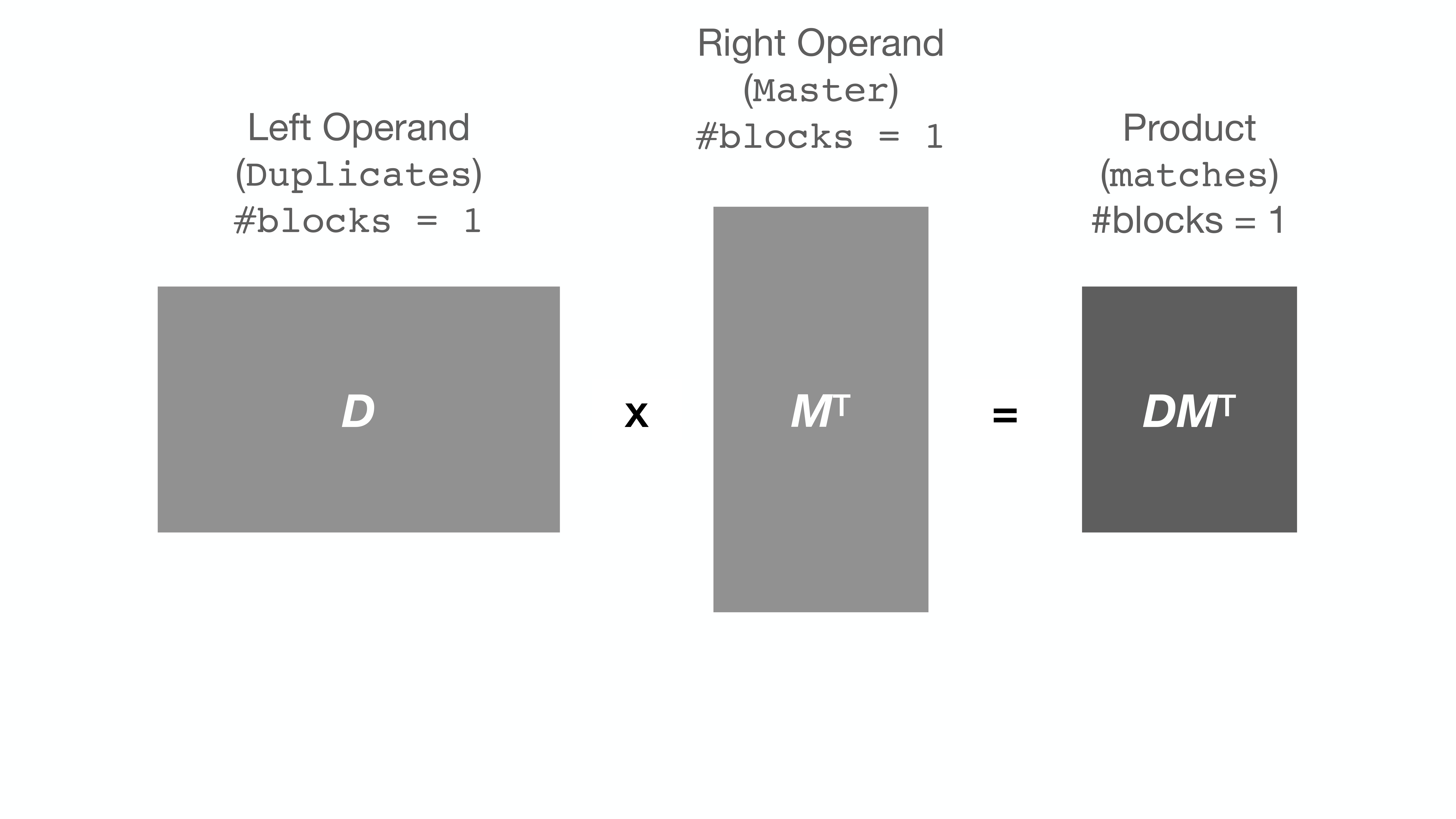
+String comparison, as implemented by `string_grouper`, is essentially matrix
+multiplication. A pandas Series of strings is converted (tokenized) into a
+matrix. Then that matrix is multiplied by itself (or another) transposed.
+
+Here is an illustration of multiplication of two matrices ***D*** and ***M***T:
+
+
+It turns out that when the matrix (or Series) is very large, the computer
+proceeds quite slowly with the multiplication (apparently due to the RAM being
+too full). Some computers give up with an `OverflowError`.
+
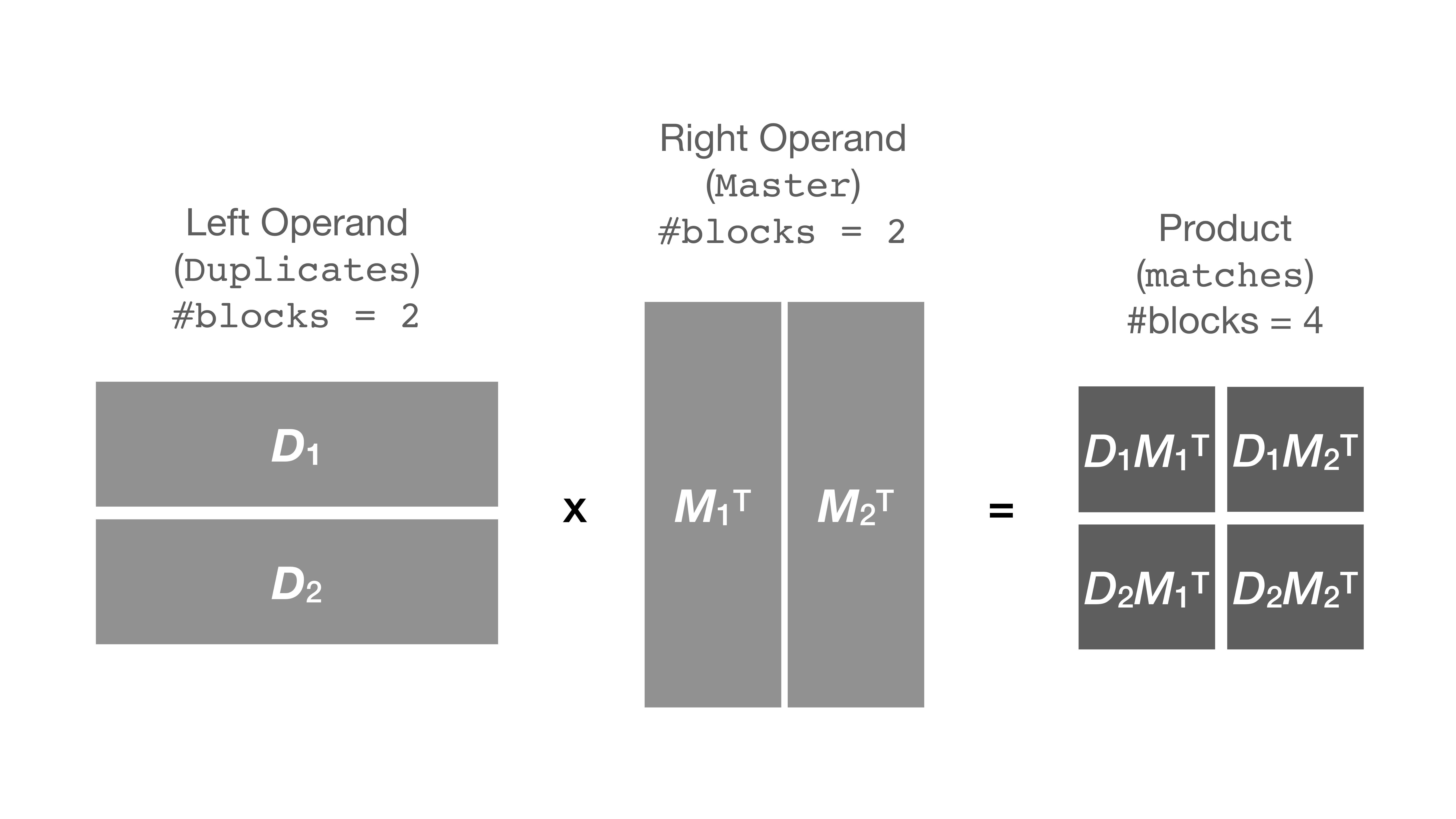
+To circumvent this issue, `string_grouper` now allows the division of the Series
+into smaller chunks (or blocks) and multiplies the chunks one pair at a time
+instead to get the same result:
+
+
+
+But surprise ... the run-time of the process is sometimes drastically reduced
+as a result. For example, the speed-up of the following call is about 500%
+(here, the Series is divided into 200 blocks on the right operand, that is,
+1 block on the left × 200 on the right) compared to the same call with no
+splitting \[`n_blocks=(1, 1)`, the default, which is what previous versions
+(0.5.0 and earlier) of `string_grouper` did\]:
+
+```python
+# A DataFrame of 668 000 records:
+companies = pd.read_csv('data/sec__edgar_company_info.csv')
+
+# The following call is more than 6 times faster than earlier versions of
+# match_strings() (that is, when n_blocks=(1, 1))!
+match_strings(companies['Company Name')], n_blocks=(1, 200))
+```
+
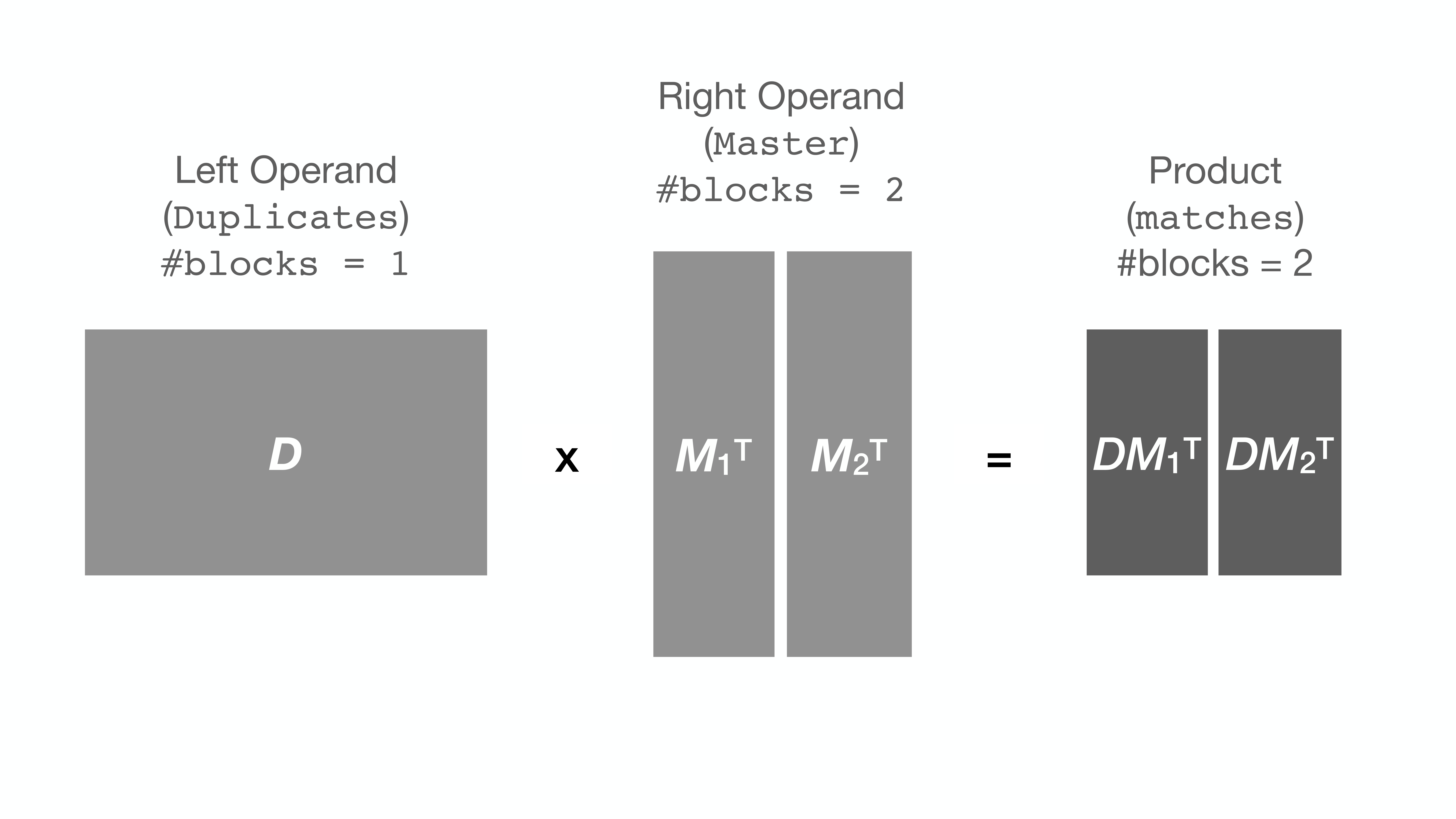
+Further exploration of the block number space ([see plot above](#Semilogx)) has revealed that for any fixed
+number of right blocks, the run-time gets longer the larger the number of left
+blocks specified. For this reason, it is recommended *not* to split the left matrix.
+
+
+
+In general,
+
+ ***total runtime*** = `n_blocks[0]` × `n_blocks[1]` × ***mean runtime per block-pair***
+
+ = ***Left Operand Size*** × ***Right Operand Size*** ×
+
+ ***mean runtime per block-pair*** / (***Left Block Size*** × ***Right Block Size***)
+
+So for given left and right operands, minimizing the ***total runtime*** is the same as minimizing the
+
+ ***runtime per string-pair comparison*** ≝
***mean runtime per block-pair*** / (***Left Block Size*** × ***Right Block Size***)
+
+
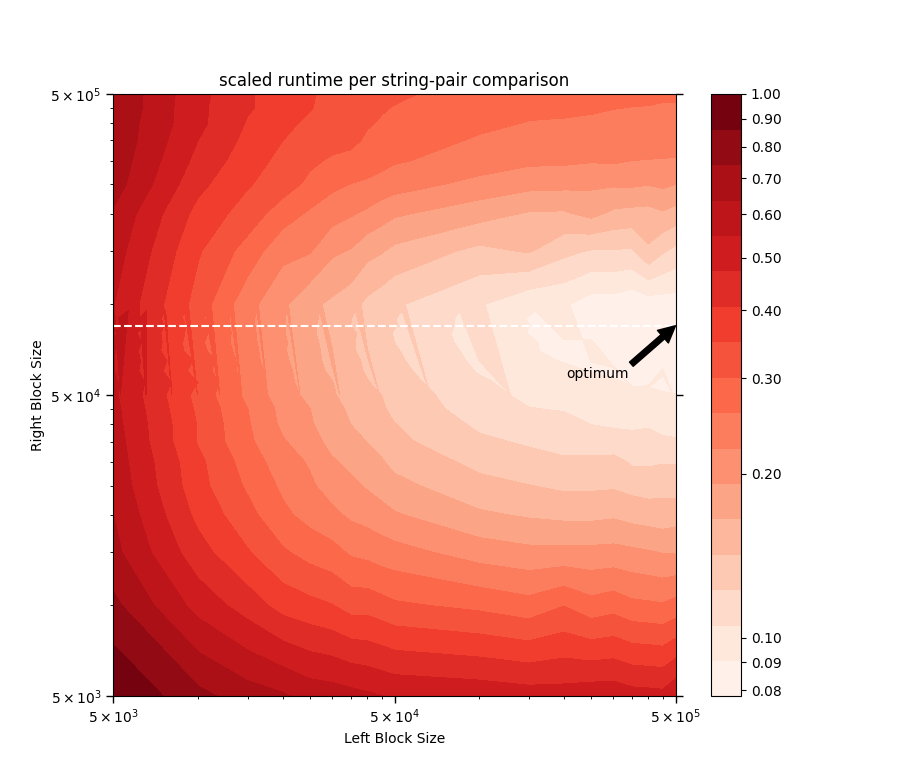
+[Below is a log-log-log contour plot](#ContourPlot) of the ***runtime per string-pair comparison*** scaled by its value
+at ***Left Block Size*** = ***Right Block Size*** = 5000. Here, ***Block Size***
+is the number of strings in that block, and ***mean runtime per block-pair*** is the time taken for the following call to run:
+```python
+# note the parameter order!
+match_strings(right_Series, left_Series, n_blocks=(1, 1))
+```
+where `left_Series` and `right_Series`, corresponding to ***Left Block*** and ***Right Block*** respectively, are random subsets of the Series `companies['Company Name')]` from the
+[sec__edgar_company_info.csv](https://www.kaggle.com/dattapiy/sec-edgar-companies-list/version/1) sample data file.
+
+ 
+
+It can be seen that when `right_Series` is roughly the size of 80 000 (denoted by the
+white dashed line in the contour plot above), the runtime per string-pair comparison is at
+its lowest for any fixed `left_Series` size. Above ***Right Block Size*** = 80 000, the
+matrix-multiplication routine begins to feel the limits of the computer's
+available memory space and thus its performance deteriorates, as evidenced by the increase
+in runtime per string-pair comparison there (above the white dashed line). This knowledge
+could serve as a guide for estimating the optimum block numbers —
+namely those that divide the Series into blocks of size roughly equal to
+80 000 for the right operand (or `right_Series`).
+
+So what are the optimum block number values for *any* given Series? That is
+anyone's guess, and may likely depend on the data itself. Furthermore, as hinted above,
+the answer may vary from computer to computer.
+
+We however encourage the user to make judicious use of the `n_blocks`
+parameter to boost performance of `string_grouper` whenever possible.
diff --git a/images/BlockMatrix_1_1.png b/images/BlockMatrix_1_1.png
new file mode 100644
index 00000000..23843452
Binary files /dev/null and b/images/BlockMatrix_1_1.png differ
diff --git a/images/BlockMatrix_1_2.png b/images/BlockMatrix_1_2.png
new file mode 100644
index 00000000..8e77511a
Binary files /dev/null and b/images/BlockMatrix_1_2.png differ
diff --git a/images/BlockMatrix_2_2.png b/images/BlockMatrix_2_2.png
new file mode 100644
index 00000000..89bbdbc5
Binary files /dev/null and b/images/BlockMatrix_2_2.png differ
diff --git a/images/BlockNumberSpaceExploration1.png b/images/BlockNumberSpaceExploration1.png
new file mode 100644
index 00000000..836600e5
Binary files /dev/null and b/images/BlockNumberSpaceExploration1.png differ
diff --git a/images/Fuzzy_vs_Exact.png b/images/Fuzzy_vs_Exact.png
new file mode 100644
index 00000000..4bfcdf39
Binary files /dev/null and b/images/Fuzzy_vs_Exact.png differ
diff --git a/images/ScaledRuntimeContourPlot.png b/images/ScaledRuntimeContourPlot.png
new file mode 100644
index 00000000..c51cea55
Binary files /dev/null and b/images/ScaledRuntimeContourPlot.png differ
diff --git a/images/ScaledTimePerComparison.png b/images/ScaledTimePerComparison.png
new file mode 100644
index 00000000..2436f54b
Binary files /dev/null and b/images/ScaledTimePerComparison.png differ
diff --git a/setup.py b/setup.py
index f4b5ecb0..cad6d08a 100644
--- a/setup.py
+++ b/setup.py
@@ -9,8 +9,8 @@
setup(
name='string_grouper',
- version='0.4.0',
- packages=['string_grouper'],
+ version='0.6.0',
+ packages=['string_grouper', 'string_grouper_utils'],
license='MIT License',
description='String grouper contains functions to do string matching using TF-IDF and the cossine similarity. '
'Based on https://bergvca.github.io/2017/10/14/super-fast-string-matching.html',
@@ -25,6 +25,7 @@
, 'scipy'
, 'scikit-learn'
, 'numpy'
- , 'sparse_dot_topn>=0.2.6'
+ , 'sparse_dot_topn_for_blocks>=0.3.1'
+ , 'topn>=0.0.7'
]
)
diff --git a/string_grouper/__init__.py b/string_grouper/__init__.py
index 84e3abd8..3b872b9b 100644
--- a/string_grouper/__init__.py
+++ b/string_grouper/__init__.py
@@ -1,2 +1,2 @@
from .string_grouper import compute_pairwise_similarities, group_similar_strings, match_most_similar, match_strings, \
-StringGrouperConfig, StringGrouper
+ StringGrouperConfig, StringGrouper
diff --git a/string_grouper/string_grouper.py b/string_grouper/string_grouper.py
index 3ab8cc46..63986354 100644
--- a/string_grouper/string_grouper.py
+++ b/string_grouper/string_grouper.py
@@ -2,15 +2,20 @@
import numpy as np
import re
import multiprocessing
+import warnings
from sklearn.feature_extraction.text import TfidfVectorizer
+from scipy.sparse import vstack
from scipy.sparse.csr import csr_matrix
+from scipy.sparse.lil import lil_matrix
from scipy.sparse.csgraph import connected_components
from typing import Tuple, NamedTuple, List, Optional, Union
-from sparse_dot_topn import awesome_cossim_topn
+from sparse_dot_topn_for_blocks import awesome_cossim_topn
+from topn import awesome_hstack_topn
from functools import wraps
-import warnings
+
DEFAULT_NGRAM_SIZE: int = 3
+DEFAULT_TFIDF_MATRIX_DTYPE: type = np.float32 # (only types np.float32 and np.float64 are allowed by sparse_dot_topn)
DEFAULT_REGEX: str = r'[,-./]|\s'
DEFAULT_MAX_N_MATCHES: int = 20
DEFAULT_MIN_SIMILARITY: float = 0.8 # minimum cosine similarity for an item to be considered a match
@@ -18,29 +23,31 @@
DEFAULT_IGNORE_CASE: bool = True # ignores case by default
DEFAULT_DROP_INDEX: bool = False # includes index-columns in output
DEFAULT_REPLACE_NA: bool = False # when finding the most similar strings, does not replace NaN values in most
- # similar string index-columns with corresponding duplicates-index values
-DEFAULT_INCLUDE_ZEROES: bool = True # when the minimum cosine similarity <=0, determines whether zero-similarity
- # matches appear in the output
-DEFAULT_SUPPRESS_WARNING: bool = False # when the minimum cosine similarity <=0 and zero-similarity matches are
- # requested, determines whether or not to suppress the message warning that
- # max_n_matches may be too small
+# similar string index-columns with corresponding duplicates-index values
+DEFAULT_INCLUDE_ZEROES: bool = True # when the minimum cosine similarity <=0, determines whether zero-similarity
+# matches appear in the output
GROUP_REP_CENTROID: str = 'centroid' # Option value to select the string in each group with the largest
- # similarity aggregate as group-representative:
+# similarity aggregate as group-representative:
GROUP_REP_FIRST: str = 'first' # Option value to select the first string in each group as group-representative:
-DEFAULT_GROUP_REP: str = GROUP_REP_CENTROID # chooses group centroid as group-representative by default
+DEFAULT_GROUP_REP: str = GROUP_REP_CENTROID # chooses group centroid as group-representative by default
+DEFAULT_FORCE_SYMMETRIES: bool = True # Option value to specify whether corrections should be made to the results
+# to account for symmetry thus compensating for those numerical errors that violate symmetry due to loss of
+# significance
+DEFAULT_N_BLOCKS: Tuple[int, int] = None # Option value to use to split dataset(s) into roughly equal-sized blocks
# The following string constants are used by (but aren't [yet] options passed to) StringGrouper
DEFAULT_COLUMN_NAME: str = 'side' # used to name non-index columns of the output of StringGrouper.get_matches
-DEFAULT_ID_NAME: str = 'id' # used to name id-columns in the output of StringGrouper.get_matches
+DEFAULT_ID_NAME: str = 'id' # used to name id-columns in the output of StringGrouper.get_matches
LEFT_PREFIX: str = 'left_' # used to prefix columns on the left of the output of StringGrouper.get_matches
RIGHT_PREFIX: str = 'right_' # used to prefix columns on the right of the output of StringGrouper.get_matches
MOST_SIMILAR_PREFIX: str = 'most_similar_' # used to prefix columns of the output of
- # StringGrouper._get_nearest_matches
-DEFAULT_MASTER_NAME: str = 'master' # used to name non-index column of the output of StringGrouper.get_nearest_matches
+# StringGrouper._get_nearest_matches
+DEFAULT_MASTER_NAME: str = 'master' # used to name non-index column of the output of StringGrouper.get_nearest_matches
DEFAULT_MASTER_ID_NAME: str = f'{DEFAULT_MASTER_NAME}_{DEFAULT_ID_NAME}' # used to name id-column of the output of
- # StringGrouper.get_nearest_matches
+# StringGrouper.get_nearest_matches
GROUP_REP_PREFIX: str = 'group_rep_' # used to prefix and name columns of the output of StringGrouper._deduplicate
+
# High level functions
@@ -55,7 +62,8 @@ def compute_pairwise_similarities(string_series_1: pd.Series,
:param kwargs: All other keyword arguments are passed to StringGrouperConfig
:return: pandas.Series of similarity scores, the same length as string_series_1 and string_series_2
"""
- return StringGrouper(string_series_1, string_series_2, **kwargs).dot()
+ sg = StringGrouper(string_series_1, string_series_2, **kwargs)
+ return sg.dot()
def group_similar_strings(strings_to_group: pd.Series,
@@ -76,8 +84,11 @@ def group_similar_strings(strings_to_group: pd.Series,
:param kwargs: All other keyword arguments are passed to StringGrouperConfig. (Optional)
:return: pandas.Series or pandas.DataFrame.
"""
- string_grouper = StringGrouper(strings_to_group, master_id=string_ids, **kwargs).fit()
- return string_grouper.get_groups()
+ sg = StringGrouper(strings_to_group,
+ master_id=string_ids,
+ **kwargs)
+ sg = sg.fit()
+ return sg.get_groups()
def match_most_similar(master: pd.Series,
@@ -105,12 +116,14 @@ def match_most_similar(master: pd.Series,
:param kwargs: All other keyword arguments are passed to StringGrouperConfig. (Optional)
:return: pandas.Series or pandas.DataFrame.
"""
- string_grouper = StringGrouper(master,
- duplicates=duplicates,
- master_id=master_id,
- duplicates_id=duplicates_id,
- **kwargs).fit()
- return string_grouper.get_groups()
+ kwargs['max_n_matches'] = 1
+ sg = StringGrouper(master,
+ duplicates=duplicates,
+ master_id=master_id,
+ duplicates_id=duplicates_id,
+ **kwargs)
+ sg = sg.fit()
+ return sg.get_groups()
def match_strings(master: pd.Series,
@@ -130,48 +143,61 @@ def match_strings(master: pd.Series,
:param kwargs: All other keyword arguments are passed to StringGrouperConfig.
:return: pandas.Dataframe.
"""
- string_grouper = StringGrouper(master,
- duplicates=duplicates,
- master_id=master_id,
- duplicates_id=duplicates_id,
- **kwargs).fit()
- return string_grouper.get_matches()
+ sg = StringGrouper(master,
+ duplicates=duplicates,
+ master_id=master_id,
+ duplicates_id=duplicates_id,
+ **kwargs)
+ sg = sg.fit()
+ return sg.get_matches()
class StringGrouperConfig(NamedTuple):
- """
+ r"""
Class with configuration variables.
:param ngram_size: int. The amount of characters in each n-gram. Default is 3.
- :param regex: str. The regex string used to cleanup the input string. Default is [,-./]|\s.
- :param max_n_matches: int. The maximum number of matches allowed per string. Default is 20.
+ :param tfidf_matrix_dtype: type. The datatype for the tf-idf values of the matrix components.
+ Possible values allowed by sparse_dot_topn are np.float32 and np.float64. Default is np.float32.
+ (Note: np.float32 often leads to faster processing and a smaller memory footprint albeit less precision
+ than np.float64.)
+ :param regex: str. The regex string used to cleanup the input string. Default is '[,-./]|\s'.
+ :param max_n_matches: int. The maximum number of matching strings in master allowed per string in duplicates.
+ Default is the total number of strings in master.
:param min_similarity: float. The minimum cosine similarity for two strings to be considered a match.
Defaults to 0.8.
:param number_of_processes: int. The number of processes used by the cosine similarity calculation.
Defaults to number of cores on a machine - 1.
:param ignore_case: bool. Whether or not case should be ignored. Defaults to True (ignore case).
:param ignore_index: whether or not to exclude string Series index-columns in output. Defaults to False.
- :param include_zeroes: when the minimum cosine similarity <=0, determines whether zero-similarity matches
+ :param include_zeroes: when the minimum cosine similarity <=0, determines whether zero-similarity matches
appear in the output. Defaults to True.
- :param suppress_warning: when min_similarity <=0 and include_zeroes=True, determines whether or not to supress
- the message warning that max_n_matches may be too small. Defaults to False.
- :param replace_na: whether or not to replace NaN values in most similar string index-columns with
+ :param replace_na: whether or not to replace NaN values in most similar string index-columns with
corresponding duplicates-index values. Defaults to False.
:param group_rep: str. The scheme to select the group-representative. Default is 'centroid'.
The other choice is 'first'.
+ :param force_symmetries: bool. In cases where duplicates is None, specifies whether corrections should be
+ made to the results to account for symmetry, thus compensating for those losses of numerical significance
+ which violate the symmetries. Defaults to True.
+ :param n_blocks: (int, int) This parameter is provided to help boost performance, if possible, of
+ processing large DataFrames, by splitting the DataFrames into n_blocks[0] blocks for the left
+ operand (of the underlying matrix multiplication) and into n_blocks[1] blocks for the right operand
+ before performing the string-comparisons block-wise. Defaults to None.
"""
ngram_size: int = DEFAULT_NGRAM_SIZE
+ tfidf_matrix_dtype: int = DEFAULT_TFIDF_MATRIX_DTYPE
regex: str = DEFAULT_REGEX
- max_n_matches: int = DEFAULT_MAX_N_MATCHES
+ max_n_matches: Optional[int] = None
min_similarity: float = DEFAULT_MIN_SIMILARITY
number_of_processes: int = DEFAULT_N_PROCESSES
ignore_case: bool = DEFAULT_IGNORE_CASE
ignore_index: bool = DEFAULT_DROP_INDEX
include_zeroes: bool = DEFAULT_INCLUDE_ZEROES
- suppress_warning: bool = DEFAULT_SUPPRESS_WARNING
replace_na: bool = DEFAULT_REPLACE_NA
group_rep: str = DEFAULT_GROUP_REP
+ force_symmetries: bool = DEFAULT_FORCE_SYMMETRIES
+ n_blocks: Tuple[int, int] = DEFAULT_N_BLOCKS
def validate_is_fit(f):
@@ -212,26 +238,130 @@ def __init__(self, master: pd.Series,
:param duplicates_id: pandas.Series. If set, contains ID values for each row in duplicates Series.
:param kwargs: All other keyword arguments are passed to StringGrouperConfig
"""
- # Validate match strings input
- if not StringGrouper._is_series_of_strings(master) or \
- (duplicates is not None and not StringGrouper._is_series_of_strings(duplicates)):
- raise TypeError('Input does not consist of pandas.Series containing only Strings')
+ # private members:
+ self.is_build = False
+
+ self._master: pd.DataFrame = pd.DataFrame()
+ self._duplicates: Optional[pd.Series] = None
+ self._master_id: Optional[pd.Series] = None
+ self._duplicates_id: Optional[pd.Series] = None
+
+ self._right_Series: pd.DataFrame = pd.DataFrame()
+ self._left_Series: pd.DataFrame = pd.DataFrame()
+
+ # After the StringGrouper is fit, _matches_list will contain the indices and similarities of the matches
+ self._matches_list: pd.DataFrame = pd.DataFrame()
+ # _true_max_n_matches will contain the true maximum number of matches over all strings in master if
+ # self._config.min_similarity <= 0
+ self._true_max_n_matches: int = 0
+ self._max_n_matches: int = 0
+
+ self._config: StringGrouperConfig = StringGrouperConfig(**kwargs)
+
+ # initialize the members:
+ self._set_data(master, duplicates, master_id, duplicates_id)
+ self._set_options(**kwargs)
+ self._build_corpus()
+
+ def _set_data(self,
+ master: pd.Series,
+ duplicates: Optional[pd.Series] = None,
+ master_id: Optional[pd.Series] = None,
+ duplicates_id: Optional[pd.Series] = None):
+ # Validate input strings data
+ self.master = master
+ self.duplicates = duplicates
+
# Validate optional IDs input
if not StringGrouper._is_input_data_combination_valid(duplicates, master_id, duplicates_id):
raise Exception('List of data Series options is invalid')
StringGrouper._validate_id_data(master, duplicates, master_id, duplicates_id)
+ self._master_id = master_id
+ self._duplicates_id = duplicates_id
+
+ # Set some private members
+ self._right_Series = self._master
+ if self._duplicates is None:
+ self._left_Series = self._master
+ else:
+ self._left_Series = self._duplicates
+
+ self.is_build = False
+
+ def _set_options(self, **kwargs):
+ self._config = StringGrouperConfig(**kwargs)
+
+ if self._config.max_n_matches is None:
+ self._max_n_matches = len(self._master)
+ else:
+ self._max_n_matches = self._config.max_n_matches
- self._master: pd.Series = master
- self._duplicates: pd.Series = duplicates if duplicates is not None else None
- self._master_id: pd.Series = master_id if master_id is not None else None
- self._duplicates_id: pd.Series = duplicates_id if duplicates_id is not None else None
- self._config: StringGrouperConfig = StringGrouperConfig(**kwargs)
self._validate_group_rep_specs()
+ self._validate_tfidf_matrix_dtype()
self._validate_replace_na_and_drop()
+ StringGrouper._validate_n_blocks(self._config.n_blocks)
+ self.is_build = False
+
+ def _build_corpus(self):
+ self._vectorizer = TfidfVectorizer(min_df=1, analyzer=self.n_grams, dtype=self._config.tfidf_matrix_dtype)
+ self._vectorizer = self._fit_vectorizer()
self.is_build = False # indicates if the grouper was fit or not
- self._vectorizer = TfidfVectorizer(min_df=1, analyzer=self.n_grams)
- # After the StringGrouper is build, _matches_list will contain the indices and similarities of two matches
- self._matches_list: pd.DataFrame = pd.DataFrame()
+
+ def reset_data(self,
+ master: pd.Series,
+ duplicates: Optional[pd.Series] = None,
+ master_id: Optional[pd.Series] = None,
+ duplicates_id: Optional[pd.Series] = None):
+ """
+ Sets the input Series of a StringGrouper instance without changing the underlying corpus.
+ :param master: pandas.Series. A Series of strings in which similar strings are searched, either against itself
+ or against the `duplicates` Series.
+ :param duplicates: pandas.Series. If set, for each string in duplicates a similar string is searched in Master.
+ :param master_id: pandas.Series. If set, contains ID values for each row in master Series.
+ :param duplicates_id: pandas.Series. If set, contains ID values for each row in duplicates Series.
+ :param kwargs: All other keyword arguments are passed to StringGrouperConfig
+ """
+ self._set_data(master, duplicates, master_id, duplicates_id)
+
+ def clear_data(self):
+ self._master = None
+ self._duplicates = None
+ self._master_id = None
+ self._duplicates_id = None
+ self._matches_list = None
+ self._left_Series = None
+ self._right_Series = None
+ self.is_build = False
+
+ def update_options(self, **kwargs):
+ """
+ Updates the kwargs of a StringGrouper object
+ :param **kwargs: any StringGrouper keyword=value argument pairs
+ """
+ _ = StringGrouperConfig(**kwargs)
+ old_kwargs = self._config._asdict()
+ old_kwargs.update(kwargs)
+ self._set_options(**old_kwargs)
+
+ @property
+ def master(self):
+ return self._master
+
+ @master.setter
+ def master(self, master):
+ if not StringGrouper._is_series_of_strings(master):
+ raise TypeError('Master input does not consist of pandas.Series containing only Strings')
+ self._master = master
+
+ @property
+ def duplicates(self):
+ return self._duplicates
+
+ @duplicates.setter
+ def duplicates(self, duplicates):
+ if duplicates is not None and not StringGrouper._is_series_of_strings(duplicates):
+ raise TypeError('Duplicates input does not consist of pandas.Series containing only Strings')
+ self._duplicates = duplicates
def n_grams(self, string: str) -> List[str]:
"""
@@ -246,16 +376,210 @@ def n_grams(self, string: str) -> List[str]:
n_grams = zip(*[string[i:] for i in range(ngram_size)])
return [''.join(n_gram) for n_gram in n_grams]
- def fit(self) -> 'StringGrouper':
- """Builds the _matches list which contains string matches indices and similarity"""
- master_matrix, duplicate_matrix = self._get_tf_idf_matrices()
- # Calculate the matches using the cosine similarity
- matches = self._build_matches(master_matrix, duplicate_matrix)
- # retrieve all matches
+ def _fit_blockwise_manual(self, n_blocks=(1, 1)):
+ # Function to compute matrix product by optionally first dividing
+ # the DataFrames(s) into equal-sized blocks as much as possible.
+
+ def divide_by(n, series):
+ # Returns an array of n rows and 2 columns.
+ # The columns denote the start and end of each of the n blocks.
+ # Note: zero-indexing is implied.
+ sz = len(series)//n
+ block_rem = np.full(n, 0, dtype=np.int64)
+ block_rem[:len(series) % n] = 1
+ if sz > 0:
+ equal_block_sz = np.full(n, sz, dtype=np.int64)
+ equal_block_sz += block_rem
+ else:
+ equal_block_sz = block_rem[:len(series) % n]
+ equal_block_sz = np.cumsum(equal_block_sz)
+ equal_block_sz = np.tile(equal_block_sz, (2, 1))
+ equal_block_sz[0, 0] = 0
+ equal_block_sz[0, 1:] = equal_block_sz[1, :-1]
+ return equal_block_sz.T
+
+ block_ranges_left = divide_by(n_blocks[0], self._left_Series)
+ block_ranges_right = divide_by(n_blocks[1], self._right_Series)
+
+ self._true_max_n_matches = 0
+ block_true_max_n_matches = 0
+ vblocks = []
+ for left_block in block_ranges_left:
+ left_matrix = self._get_left_tf_idf_matrix(left_block)
+ nnz_rows = np.full(left_matrix.shape[0], 0, dtype=np.int32)
+ hblocks = []
+ for right_block in block_ranges_right:
+ right_matrix = self._get_right_tf_idf_matrix(right_block)
+ try:
+ # Calculate the matches using the cosine similarity
+ # Note: awesome_cossim_topn will sort each row only when
+ # _max_n_matches < size of right_block or sort=True
+ matches, block_true_max_n_matches = self._build_matches(
+ left_matrix, right_matrix, nnz_rows, sort=(len(block_ranges_right) == 1)
+ )
+ except OverflowError as oe:
+ import sys
+ raise (type(oe)(f"{str(oe)} Use the n_blocks parameter to split-up "
+ f"the data into smaller chunks. The current values"
+ f"(n_blocks = {n_blocks}) are too small.")
+ .with_traceback(sys.exc_info()[2]))
+ hblocks.append(matches)
+ # end of inner loop
+
+ self._true_max_n_matches = \
+ max(block_true_max_n_matches, self._true_max_n_matches)
+ if len(block_ranges_right) > 1:
+ # Note: awesome_hstack_topn will sort each row only when
+ # _max_n_matches < length of _right_Series or sort=True
+ vblocks.append(
+ awesome_hstack_topn(
+ hblocks,
+ self._max_n_matches,
+ sort=True,
+ use_threads=self._config.number_of_processes > 1,
+ n_jobs=self._config.number_of_processes
+ )
+ )
+ else:
+ vblocks.append(hblocks[0])
+ del hblocks
+ del matches
+ # end of outer loop
+
+ if len(block_ranges_left) > 1:
+ return vstack(vblocks)
+ else:
+ return vblocks[0]
+
+ def _fit_blockwise_auto(self,
+ left_partition=(None, None),
+ right_partition=(None, None),
+ nnz_rows=None,
+ sort=True,
+ whoami=0):
+ # This is a recursive function!
+ # fit() has been extended here to enable StringGrouper to handle large
+ # datasets which otherwise would lead to an OverflowError
+ # The handling is achieved using block matrix multiplication.
+ def begin(partition):
+ return partition[0] if partition[0] is not None else 0
+
+ def end(partition, left=True):
+ if partition[1] is not None:
+ return partition[1]
+
+ return len(self._left_Series if left else self._right_Series)
+
+ left_matrix = self._get_left_tf_idf_matrix(left_partition)
+ right_matrix = self._get_right_tf_idf_matrix(right_partition)
+
+ if whoami == 0:
+ # At the topmost level of recursion initialize nnz_rows
+ # which will be used to compute _true_max_n_matches
+ nnz_rows = np.full(left_matrix.shape[0], 0, dtype=np.int32)
+ self._true_max_n_matches = 0
+
+ try:
+ # Calculate the matches using the cosine similarity
+ matches, true_max_n_matches = self._build_matches(
+ left_matrix, right_matrix, nnz_rows[slice(*left_partition)],
+ sort=sort)
+ except OverflowError:
+ warnings.warn("An OverflowError occurred but is being "
+ "handled. The input data will be automatically "
+ "split-up into smaller chunks which will then be "
+ "processed one chunk at a time. To prevent "
+ "OverflowError, use the n_blocks parameter to split-up "
+ "the data manually into small enough chunks.")
+ # Matrices too big! Try splitting:
+ del left_matrix, right_matrix
+
+ def split_partition(partition, left=True):
+ data_begin = begin(partition)
+ data_end = end(partition, left=left)
+ data_mid = data_begin + (data_end - data_begin)//2
+ if data_mid > data_begin:
+ return [(data_begin, data_mid), (data_mid, data_end)]
+ else:
+ return [(data_begin, data_end)]
+
+ left_halves = split_partition(left_partition, left=True)
+ right_halves = split_partition(right_partition, left=False)
+ vblocks = []
+ for lhalf in left_halves:
+ hblocks = []
+ for rhalf in right_halves:
+ # Note: awesome_cossim_topn will sort each row only when
+ # _max_n_matches < size of right_partition or sort=True
+ matches = self._fit_blockwise_auto(
+ left_partition=lhalf, right_partition=rhalf,
+ nnz_rows=nnz_rows,
+ sort=((whoami == 0) and (len(right_halves) == 1)),
+ whoami=(whoami + 1)
+ )
+ hblocks.append(matches)
+ # end of inner loop
+ if whoami == 0:
+ self._true_max_n_matches = max(
+ np.amax(nnz_rows[slice(*lhalf)]),
+ self._true_max_n_matches
+ )
+ if len(right_halves) > 1:
+ # Note: awesome_hstack_topn will sort each row only when
+ # _max_n_matches < length of _right_Series or sort=True
+ vblocks.append(
+ awesome_hstack_topn(
+ hblocks,
+ self._max_n_matches,
+ sort=(whoami == 0),
+ use_threads=self._config.number_of_processes > 1,
+ n_jobs=self._config.number_of_processes
+ )
+ )
+ else:
+ vblocks.append(hblocks[0])
+ del hblocks
+ # end of outer loop
+ if len(left_halves) > 1:
+ return vstack(vblocks)
+ else:
+ return vblocks[0]
+
+ if whoami == 0:
+ self._true_max_n_matches = true_max_n_matches
+ return matches
+
+ def fit(self, force_symmetries=None, n_blocks=None):
+ """
+ Builds the _matches list which contains string-matches' indices and similarity
+ Updates and returns the StringGrouper object that called it.
+ """
+ if force_symmetries is None:
+ force_symmetries = self._config.force_symmetries
+ StringGrouper._validate_n_blocks(n_blocks)
+ if n_blocks is None:
+ n_blocks = self._config.n_blocks
+
+ # do the matching
+ if n_blocks is None:
+ matches = self._fit_blockwise_auto()
+ else:
+ matches = self._fit_blockwise_manual(n_blocks=n_blocks)
+
+ # enforce symmetries?
+ if force_symmetries and (self._duplicates is None):
+ # convert to lil format for best efficiency when setting
+ # matrix-elements
+ matches = matches.tolil()
+ # matrix diagonal elements must be exactly 1 (numerical precision
+ # errors introduced by floating-point computations in
+ # awesome_cossim_topn sometimes lead to unexpected results)
+ matches = StringGrouper._fix_diagonal(matches)
+ # the list of matches must be symmetric!
+ # (i.e., if A != B and A matches B; then B matches A)
+ matches = StringGrouper._symmetrize_matrix(matches)
+ matches = matches.tocsr()
self._matches_list = self._get_matches_list(matches)
- if self._duplicates is None:
- # the list of matches needs to be symmetric!!! (i.e., if A != B and A matches B; then B matches A)
- self._symmetrize_matches_list()
self.is_build = True
return self
@@ -263,26 +587,23 @@ def dot(self) -> pd.Series:
"""Computes the row-wise similarity scores between strings in _master and _duplicates"""
if len(self._master) != len(self._duplicates):
raise Exception("To perform this function, both input Series must have the same length.")
- master_matrix, duplicate_matrix = self._get_tf_idf_matrices()
+ master_matrix, duplicate_matrix = self._get_left_tf_idf_matrix(), self._get_right_tf_idf_matrix()
# Calculate pairwise cosine similarities:
- pairwise_similarities = np.asarray(master_matrix.multiply(duplicate_matrix).sum(axis=1)).squeeze()
+ pairwise_similarities = np.asarray(master_matrix.multiply(duplicate_matrix).sum(axis=1)).squeeze(axis=1)
return pd.Series(pairwise_similarities, name='similarity', index=self._master.index)
@validate_is_fit
def get_matches(self,
ignore_index: Optional[bool] = None,
- include_zeroes: Optional[bool]=None,
- suppress_warning: Optional[bool]=None) -> pd.DataFrame:
+ include_zeroes: Optional[bool] = None) -> pd.DataFrame:
"""
Returns a DataFrame with all the matches and their cosine similarity.
If optional IDs are used, returned as extra columns with IDs matched to respective data rows
- :param ignore_index: whether or not to exclude string Series index-columns in output. Defaults to
+ :param ignore_index: whether or not to exclude string Series index-columns in output. Defaults to
self._config.ignore_index.
- :param include_zeroes: when the minimum cosine similarity <=0, determines whether zero-similarity matches
+ :param include_zeroes: when the minimum cosine similarity <=0, determines whether zero-similarity matches
appear in the output. Defaults to self._config.include_zeroes.
- :param suppress_warning: when min_similarity <=0 and include_zeroes=True, determines whether or not to suppress
- the message warning that max_n_matches may be too small. Defaults to self._config.suppress_warning.
"""
def get_both_sides(master: pd.Series,
duplicates: pd.Series,
@@ -304,19 +625,20 @@ def prefix_column_names(data: Union[pd.Series, pd.DataFrame], prefix: str):
else:
return data.rename(f"{prefix}{data.name}")
- if ignore_index is None: ignore_index = self._config.ignore_index
- if include_zeroes is None: include_zeroes = self._config.include_zeroes
- if suppress_warning is None: suppress_warning = self._config.suppress_warning
+ if ignore_index is None:
+ ignore_index = self._config.ignore_index
+ if include_zeroes is None:
+ include_zeroes = self._config.include_zeroes
if self._config.min_similarity > 0 or not include_zeroes:
matches_list = self._matches_list
elif include_zeroes:
# Here's a fix to a bug pointed out by one GitHub user (@nbcvijanovic):
- # the fix includes zero-similarity matches that are missing by default
- # in _matches_list due to our use of sparse matrices
- non_matches_list = self._get_non_matches_list(suppress_warning)
+ # the fix includes zero-similarity matches that are missing by default
+ # in _matches_list due to our use of sparse matrices
+ non_matches_list = self._get_non_matches_list()
matches_list = self._matches_list if non_matches_list.empty else \
pd.concat([self._matches_list, non_matches_list], axis=0, ignore_index=True)
-
+
left_side, right_side = get_both_sides(self._master, self._duplicates, drop_index=ignore_index)
similarity = matches_list.similarity.reset_index(drop=True)
if self._master_id is None:
@@ -358,18 +680,128 @@ def get_groups(self,
If there are IDs (master_id and/or duplicates_id) then the IDs corresponding to the string outputs
above are returned as well altogether in a DataFrame.
- :param ignore_index: whether or not to exclude string Series index-columns in output. Defaults to
+ :param ignore_index: whether or not to exclude string Series index-columns in output. Defaults to
self._config.ignore_index.
- :param replace_na: whether or not to replace NaN values in most similar string index-columns with
+ :param replace_na: whether or not to replace NaN values in most similar string index-columns with
corresponding duplicates-index values. Defaults to self._config.replace_na.
"""
- if ignore_index is None: ignore_index = self._config.ignore_index
+ if ignore_index is None:
+ ignore_index = self._config.ignore_index
if self._duplicates is None:
return self._deduplicate(ignore_index=ignore_index)
else:
- if replace_na is None: replace_na = self._config.replace_na
+ if replace_na is None:
+ replace_na = self._config.replace_na
return self._get_nearest_matches(ignore_index=ignore_index, replace_na=replace_na)
+ def match_strings(self,
+ master: pd.Series,
+ duplicates: Optional[pd.Series] = None,
+ master_id: Optional[pd.Series] = None,
+ duplicates_id: Optional[pd.Series] = None,
+ **kwargs) -> pd.DataFrame:
+ """
+ Returns all highly similar strings without rebuilding the corpus.
+ If only 'master' is given, it will return highly similar strings within master.
+ This can be seen as an self-join. If both master and duplicates is given, it will return highly similar strings
+ between master and duplicates. This can be seen as an inner-join.
+
+ :param master: pandas.Series. Series of strings against which matches are calculated.
+ :param duplicates: pandas.Series. Series of strings that will be matched with master if given (Optional).
+ :param master_id: pandas.Series. Series of values that are IDs for master column rows (Optional).
+ :param duplicates_id: pandas.Series. Series of values that are IDs for duplicates column rows (Optional).
+ :param kwargs: All other keyword arguments are passed to StringGrouperConfig.
+ :return: pandas.Dataframe.
+ """
+ self.reset_data(master, duplicates, master_id, duplicates_id)
+ self.update_options(**kwargs)
+ self = self.fit()
+ return self.get_matches()
+
+ def match_most_similar(self,
+ master: pd.Series,
+ duplicates: pd.Series,
+ master_id: Optional[pd.Series] = None,
+ duplicates_id: Optional[pd.Series] = None,
+ **kwargs) -> Union[pd.DataFrame, pd.Series]:
+ """
+ If no IDs ('master_id' and 'duplicates_id') are given, returns, without rebuilding the corpus, a
+ Series of strings of the same length as 'duplicates' where for each string in duplicates the most
+ similar string in 'master' is returned.
+ If there are no similar strings in master for a given string in duplicates
+ (there is no potential match where the cosine similarity is above the threshold [default: 0.8])
+ the original string in duplicates is returned.
+
+ For example the input Series [foooo, bar, baz] (master) and [foooob, bar, new] will return:
+ [foooo, bar, new].
+
+ If IDs (both 'master_id' and 'duplicates_id') are also given, returns a DataFrame of the same strings
+ output in the above case with their corresponding IDs.
+
+ :param master: pandas.Series. Series of strings that the duplicates will be matched with.
+ :param duplicates: pandas.Series. Series of strings that will me matched with the master.
+ :param master_id: pandas.Series. Series of values that are IDs for master column rows. (Optional)
+ :param duplicates_id: pandas.Series. Series of values that are IDs for duplicates column rows. (Optional)
+ :param kwargs: All other keyword arguments are passed to StringGrouperConfig. (Optional)
+ :return: pandas.Series or pandas.DataFrame.
+ """
+ self.reset_data(master, duplicates, master_id, duplicates_id)
+
+ old_max_n_matches = self._max_n_matches
+ new_max_n_matches = None
+ if 'max_n_matches' in kwargs:
+ new_max_n_matches = kwargs['max_n_matches']
+ kwargs['max_n_matches'] = 1
+ self.update_options(**kwargs)
+
+ self = self.fit()
+ output = self.get_groups()
+
+ kwargs['max_n_matches'] = old_max_n_matches if new_max_n_matches is None else new_max_n_matches
+ self.update_options(**kwargs)
+ return output
+
+ def group_similar_strings(self,
+ strings_to_group: pd.Series,
+ string_ids: Optional[pd.Series] = None,
+ **kwargs) -> Union[pd.DataFrame, pd.Series]:
+ """
+ If 'string_ids' is not given, finds all similar strings in 'strings_to_group' without rebuilding the
+ corpus and returns a Series of strings of the same length as 'strings_to_group'. For each group of
+ similar strings a single string is chosen as the 'master' string and is returned for each member of
+ the group.
+
+ For example the input Series: [foooo, foooob, bar] will return [foooo, foooo, bar]. Here 'foooo' and
+ 'foooob' are grouped together into group 'foooo' because they are found to be very similar.
+
+ If string_ids is also given, a DataFrame of the strings and their corresponding IDs is instead returned.
+
+ :param strings_to_group: pandas.Series. The input Series of strings to be grouped.
+ :param string_ids: pandas.Series. The input Series of the IDs of the strings to be grouped. (Optional)
+ :param kwargs: All other keyword arguments are passed to StringGrouperConfig. (Optional)
+ :return: pandas.Series or pandas.DataFrame.
+ """
+ self.reset_data(strings_to_group, master_id=string_ids)
+ self.update_options(**kwargs)
+ self = self.fit()
+ return self.get_groups()
+
+ def compute_pairwise_similarities(self,
+ string_series_1: pd.Series,
+ string_series_2: pd.Series,
+ **kwargs) -> pd.Series:
+ """
+ Computes the similarity scores between two Series of strings row-wise without rebuilding the corpus.
+
+ :param string_series_1: pandas.Series. The input Series of strings to be grouped
+ :param string_series_2: pandas.Series. The input Series of the IDs of the strings to be grouped
+ :param kwargs: All other keyword arguments are passed to StringGrouperConfig
+ :return: pandas.Series of similarity scores, the same length as string_series_1 and string_series_2
+ """
+ self.reset_data(string_series_1, string_series_2)
+ self.update_options(**kwargs)
+ return self.dot()
+
@validate_is_fit
def add_match(self, master_side: str, dupe_side: str) -> 'StringGrouper':
"""Adds a match if it wasn't found by the fit function"""
@@ -409,19 +841,19 @@ def remove_match(self, master_side: str, dupe_side: str) -> 'StringGrouper':
)]
return self
- def _get_tf_idf_matrices(self) -> Tuple[csr_matrix, csr_matrix]:
- # Fit the tf-idf vectorizer
- self._vectorizer = self._fit_vectorizer()
- # Build the two matrices
- master_matrix = self._vectorizer.transform(self._master)
-
- if self._duplicates is not None:
- duplicate_matrix = self._vectorizer.transform(self._duplicates)
- # IF there is no duplicate matrix, we assume we want to match on the master matrix itself
- else:
- duplicate_matrix = master_matrix
+ def _get_left_tf_idf_matrix(self, partition=(None, None)):
+ # unlike _get_tf_idf_matrices(), _get_left_tf_idf_matrix
+ # does not set the corpus but rather
+ # builds a matrix using the existing corpus
+ return self._vectorizer.transform(
+ self._left_Series.iloc[slice(*partition)])
- return master_matrix, duplicate_matrix
+ def _get_right_tf_idf_matrix(self, partition=(None, None)):
+ # unlike _get_tf_idf_matrices(), _get_right_tf_idf_matrix
+ # does not set the corpus but rather
+ # builds a matrix using the existing corpus
+ return self._vectorizer.transform(
+ self._right_Series.iloc[slice(*partition)])
def _fit_vectorizer(self) -> TfidfVectorizer:
# if both dupes and master string series are set - we concat them to fit the vectorizer on all
@@ -433,74 +865,57 @@ def _fit_vectorizer(self) -> TfidfVectorizer:
self._vectorizer.fit(strings)
return self._vectorizer
- def _build_matches(self, master_matrix: csr_matrix, duplicate_matrix: csr_matrix) -> csr_matrix:
+ def _build_matches(self,
+ left_matrix: csr_matrix, right_matrix: csr_matrix,
+ nnz_rows: np.ndarray = None,
+ sort: bool = True) -> csr_matrix:
"""Builds the cossine similarity matrix of two csr matrices"""
- tf_idf_matrix_1 = master_matrix
- tf_idf_matrix_2 = duplicate_matrix.transpose()
-
- optional_kwargs = dict()
- if self._config.number_of_processes > 1:
- optional_kwargs = {
- 'use_threads': True,
- 'n_jobs': self._config.number_of_processes
- }
-
- return awesome_cossim_topn(tf_idf_matrix_1, tf_idf_matrix_2,
- self._config.max_n_matches,
- self._config.min_similarity,
- **optional_kwargs)
-
- def _symmetrize_matches_list(self):
- # [symmetrized matches_list] = [matches_list] UNION [transposed matches_list] (i.e., column-names swapped):
- self._matches_list = self._matches_list.set_index(['master_side', 'dupe_side'])\
- .combine_first(
- self._matches_list.rename(
- columns={
- 'master_side': 'dupe_side',
- 'dupe_side': 'master_side'
- }
- ).set_index(['master_side', 'dupe_side'])
- ).reset_index()
-
- def _get_non_matches_list(self, suppress_warning=False) -> pd.DataFrame:
+ right_matrix = right_matrix.transpose()
+
+ if nnz_rows is None:
+ nnz_rows = np.full(left_matrix.shape[0], 0, dtype=np.int32)
+
+ optional_kwargs = {
+ 'return_best_ntop': True,
+ 'sort': sort,
+ 'use_threads': self._config.number_of_processes > 1,
+ 'n_jobs': self._config.number_of_processes}
+
+ return awesome_cossim_topn(
+ left_matrix, right_matrix,
+ self._max_n_matches,
+ nnz_rows,
+ self._config.min_similarity,
+ **optional_kwargs)
+
+ def _get_matches_list(self,
+ matches: csr_matrix
+ ) -> pd.DataFrame:
+ """Returns a list of all the indices of matches"""
+ r, c = matches.nonzero()
+ d = matches.data
+ return pd.DataFrame({'master_side': c.astype(np.int64),
+ 'dupe_side': r.astype(np.int64),
+ 'similarity': d})
+
+ def _get_non_matches_list(self) -> pd.DataFrame:
"""Returns a list of all the indices of non-matching pairs (with similarity set to 0)"""
m_sz, d_sz = len(self._master), len(self._master if self._duplicates is None else self._duplicates)
all_pairs = pd.MultiIndex.from_product([range(m_sz), range(d_sz)], names=['master_side', 'dupe_side'])
matched_pairs = pd.MultiIndex.from_frame(self._matches_list[['master_side', 'dupe_side']])
missing_pairs = all_pairs.difference(matched_pairs)
- if missing_pairs.empty: return pd.DataFrame()
- if (self._config.max_n_matches < d_sz) and not suppress_warning:
- warnings.warn(f'WARNING: max_n_matches={self._config.max_n_matches} may be too small!\n'
- f'\t\t Some zero-similarity matches returned may be false!\n'
- f'\t\t To be absolutely certain all zero-similarity matches are true,\n'
- f'\t\t try setting max_n_matches={d_sz} (the length of the Series parameter duplicates).\n'
- f'\t\t To suppress this warning, set suppress_warning=True.')
+ if missing_pairs.empty:
+ return pd.DataFrame()
+ if (self._max_n_matches < self._true_max_n_matches):
+ raise Exception(f'\nERROR: Cannot return zero-similarity matches since \n'
+ f'\t\t max_n_matches={self._max_n_matches} is too small!\n'
+ f'\t\t Try setting max_n_matches={self._true_max_n_matches} (the \n'
+ f'\t\t true maximum number of matches over all strings in master)\n'
+ f'\t\t or greater or do not set this kwarg at all.')
missing_pairs = missing_pairs.to_frame(index=False)
missing_pairs['similarity'] = 0
return missing_pairs
- @staticmethod
- def _get_matches_list(matches) -> pd.DataFrame:
- """Returns a list of all the indices of matches"""
- non_zeros = matches.nonzero()
-
- sparserows = non_zeros[0]
- sparsecols = non_zeros[1]
- nr_matches = sparsecols.size
- master_side = np.empty([nr_matches], dtype=np.int64)
- dupe_side = np.empty([nr_matches], dtype=np.int64)
- similarity = np.zeros(nr_matches)
-
- for index in range(0, nr_matches):
- master_side[index] = sparserows[index]
- dupe_side[index] = sparsecols[index]
- similarity[index] = matches.data[index]
-
- matches_list = pd.DataFrame({'master_side': master_side,
- 'dupe_side': dupe_side,
- 'similarity': similarity})
- return matches_list
-
def _get_nearest_matches(self,
ignore_index=False,
replace_na=False) -> Union[pd.DataFrame, pd.Series]:
@@ -508,8 +923,8 @@ def _get_nearest_matches(self,
master_label = f'{prefix}{self._master.name if self._master.name else DEFAULT_MASTER_NAME}'
master = self._master.rename(master_label).reset_index(drop=ignore_index)
dupes = self._duplicates.rename('duplicates').reset_index(drop=ignore_index)
-
- # Rename new master-columns to avoid possible conflict with new dupes-columns when later merging
+
+ # Rename new master-columns to avoid possible conflict with new dupes-columns when later merging
if isinstance(dupes, pd.DataFrame):
master.rename(
columns={col: f'{prefix}{col}' for col in master.columns if str(col) != master_label},
@@ -539,14 +954,14 @@ def _get_nearest_matches(self,
if self._master_id is not None:
# Also update the master_id-series with the duplicates_id in cases were there is no match
dupes_max_sim.loc[rows_to_update, master_id_label] = dupes_max_sim[rows_to_update].duplicates_id
-
+
# For some weird reason, pandas' merge function changes int-datatype columns to float when NaN values
# appear within them. So here we change them back to their original datatypes if possible:
if dupes_max_sim[master_id_label].dtype != self._master_id.dtype and \
- self._duplicates_id.dtype == self._master_id.dtype:
+ self._duplicates_id.dtype == self._master_id.dtype:
dupes_max_sim.loc[:, master_id_label] = \
- dupes_max_sim.loc[:, master_id_label].astype(self._master_id.dtype)
-
+ dupes_max_sim.loc[:, master_id_label].astype(self._master_id.dtype)
+
# Prepare the output:
required_column_list = [master_label] if self._master_id is None else [master_id_label, master_label]
index_column_list = \
@@ -556,22 +971,21 @@ def _get_nearest_matches(self,
# Update the master index-columns with the duplicates index-column values in cases were there is no match
dupes_index_columns = [col for col in dupes.columns if str(col) != 'duplicates']
dupes_max_sim.loc[rows_to_update, index_column_list] = \
- dupes_max_sim.loc[rows_to_update, dupes_index_columns].values
-
+ dupes_max_sim.loc[rows_to_update, dupes_index_columns].values
+
# Restore their original datatypes if possible:
for m, d in zip(index_column_list, dupes_index_columns):
if dupes_max_sim[m].dtype != master[m].dtype and dupes[d].dtype == master[m].dtype:
dupes_max_sim.loc[:, m] = dupes_max_sim.loc[:, m].astype(master[m].dtype)
-
+
# Make sure to keep same order as duplicates
dupes_max_sim = dupes_max_sim.sort_values('dupe_side').set_index('dupe_side')
output = dupes_max_sim[index_column_list + required_column_list]
output.index = self._duplicates.index
- return output.squeeze()
+ return output.squeeze(axis=1)
def _deduplicate(self, ignore_index=False) -> Union[pd.DataFrame, pd.Series]:
- # discard self-matches: A matches A
- pairs = self._matches_list[self._matches_list['master_side'] != self._matches_list['dupe_side']]
+ pairs = self._matches_list
# rebuild graph adjacency matrix from already found matches:
n = len(self._master)
graph = csr_matrix(
@@ -599,7 +1013,7 @@ def _deduplicate(self, ignore_index=False) -> Union[pd.DataFrame, pd.Series]:
graph.data = pairs['similarity'].to_numpy()
# sum along the rows to obtain numpy 1D matrix of similarity aggregates then ...
# ... convert to 1D numpy array (using asarray then squeeze) and then to Series:
- group_of_master_index['weight'] = pd.Series(np.asarray(graph.sum(axis=1)).squeeze())
+ group_of_master_index['weight'] = pd.Series(np.asarray(graph.sum(axis=1)).squeeze(axis=1))
method = 'idxmax'
# Determine the group representatives AND merge with indices:
@@ -623,7 +1037,7 @@ def _deduplicate(self, ignore_index=False) -> Union[pd.DataFrame, pd.Series]:
output_id = self._master_id.iloc[group_of_master_index.group_rep].rename(id_label).reset_index(drop=True)
output = pd.concat([output_id, output], axis=1)
output.index = self._master.index
- return output.squeeze()
+ return output
def _get_indices_of(self, master_side: str, dupe_side: str) -> Tuple[pd.Series, pd.Series]:
master_strings = self._master
@@ -634,7 +1048,7 @@ def _get_indices_of(self, master_side: str, dupe_side: str) -> Tuple[pd.Series,
master_indices = master_strings[master_strings == master_side].index.to_series().reset_index(drop=True)
dupe_indices = dupe_strings[dupe_strings == dupe_side].index.to_series().reset_index(drop=True)
return master_indices, dupe_indices
-
+
def _validate_group_rep_specs(self):
group_rep_options = (GROUP_REP_FIRST, GROUP_REP_CENTROID)
if self._config.group_rep not in group_rep_options:
@@ -642,6 +1056,13 @@ def _validate_group_rep_specs(self):
f"Invalid option value for group_rep. The only permitted values are\n {group_rep_options}"
)
+ def _validate_tfidf_matrix_dtype(self):
+ dtype_options = (np.float32, np.float64)
+ if self._config.tfidf_matrix_dtype not in dtype_options:
+ raise Exception(
+ f"Invalid option value for tfidf_matrix_dtype. The only permitted values are\n {dtype_options}"
+ )
+
def _validate_replace_na_and_drop(self):
if self._config.ignore_index and self._config.replace_na:
raise Exception("replace_na can only be set to True when ignore_index=False.")
@@ -651,6 +1072,33 @@ def _validate_replace_na_and_drop(self):
"index if the number of index-levels does not equal the number of index-columns."
)
+ @staticmethod
+ def _validate_n_blocks(n_blocks):
+ errmsg = "Invalid option value for parameter n_blocks: "
+ "n_blocks must be None or a tuple of 2 integers greater than 0."
+ if n_blocks is None:
+ return
+ if not isinstance(n_blocks, tuple):
+ raise Exception(errmsg)
+ if len(n_blocks) != 2:
+ raise Exception(errmsg)
+ if not (isinstance(n_blocks[0], int) and isinstance(n_blocks[1], int)):
+ raise Exception(errmsg)
+ if (n_blocks[0] < 1) or (n_blocks[1] < 1):
+ raise Exception(errmsg)
+
+ @staticmethod
+ def _fix_diagonal(m: lil_matrix) -> lil_matrix:
+ r = np.arange(m.shape[0])
+ m[r, r] = 1
+ return m
+
+ @staticmethod
+ def _symmetrize_matrix(m_symmetric: lil_matrix) -> lil_matrix:
+ r, c = m_symmetric.nonzero()
+ m_symmetric[c, r] = m_symmetric[r, c]
+ return m_symmetric
+
@staticmethod
def _make_symmetric(new_matches: pd.DataFrame) -> pd.DataFrame:
columns_switched = pd.DataFrame({'master_side': new_matches.dupe_side,
@@ -678,7 +1126,7 @@ def _is_series_of_strings(series_to_test: pd.Series) -> bool:
return False
elif series_to_test.to_frame().applymap(
lambda x: not isinstance(x, str)
- ).squeeze().any():
+ ).squeeze(axis=1).any():
return False
return True
diff --git a/string_grouper/test/test_string_grouper.py b/string_grouper/test/test_string_grouper.py
index 723d3f22..b159646b 100644
--- a/string_grouper/test/test_string_grouper.py
+++ b/string_grouper/test/test_string_grouper.py
@@ -3,13 +3,15 @@
import numpy as np
from scipy.sparse.csr import csr_matrix
from string_grouper.string_grouper import DEFAULT_MIN_SIMILARITY, \
- DEFAULT_MAX_N_MATCHES, DEFAULT_REGEX, \
- DEFAULT_NGRAM_SIZE, DEFAULT_N_PROCESSES, DEFAULT_IGNORE_CASE, \
+ DEFAULT_REGEX, DEFAULT_NGRAM_SIZE, DEFAULT_N_PROCESSES, DEFAULT_IGNORE_CASE, \
StringGrouperConfig, StringGrouper, StringGrouperNotFitException, \
- match_most_similar, group_similar_strings, match_strings,\
+ match_most_similar, group_similar_strings, match_strings, \
compute_pairwise_similarities
-from unittest.mock import patch
-import warnings
+from unittest.mock import patch, Mock
+
+
+def mock_symmetrize_matrix(x: csr_matrix) -> csr_matrix:
+ return x
class SimpleExample(object):
@@ -93,7 +95,7 @@ def test_config_defaults(self):
"""Empty initialisation should set default values"""
config = StringGrouperConfig()
self.assertEqual(config.min_similarity, DEFAULT_MIN_SIMILARITY)
- self.assertEqual(config.max_n_matches, DEFAULT_MAX_N_MATCHES)
+ self.assertEqual(config.max_n_matches, None)
self.assertEqual(config.regex, DEFAULT_REGEX)
self.assertEqual(config.ngram_size, DEFAULT_NGRAM_SIZE)
self.assertEqual(config.number_of_processes, DEFAULT_N_PROCESSES)
@@ -114,6 +116,251 @@ def test_config_non_default_values(self):
class StringGrouperTest(unittest.TestCase):
+
+ def test_auto_blocking_single_DataFrame(self):
+ """tests whether automatic blocking yields consistent results"""
+ # This function will force an OverflowError to occur when
+ # the input Series have a combined length above a given number:
+ # OverflowThreshold. This will in turn trigger automatic splitting
+ # of the Series/matrices into smaller blocks when n_blocks = None
+
+ sort_cols = ['right_index', 'left_index']
+
+ def fix_row_order(df):
+ return df.sort_values(sort_cols).reset_index(drop=True)
+
+ simple_example = SimpleExample()
+ df1 = simple_example.customers_df2['Customer Name']
+
+ # first do manual blocking
+ sg = StringGrouper(df1, min_similarity=0.1)
+ pd.testing.assert_series_equal(sg.master, df1)
+ self.assertEqual(sg.duplicates, None)
+
+ matches = fix_row_order(sg.match_strings(df1, n_blocks=(1, 1)))
+ self.assertEqual(sg._config.n_blocks, (1, 1))
+
+ # Create a custom wrapper for this StringGrouper instance's
+ # _build_matches() method which will later be used to
+ # mock _build_matches().
+ # Note that we have to define the wrapper here because
+ # _build_matches() is a non-static function of StringGrouper
+ # and needs access to the specific StringGrouper instance sg
+ # created here.
+ def mock_build_matches(OverflowThreshold,
+ real_build_matches=sg._build_matches):
+ def wrapper(left_matrix,
+ right_matrix,
+ nnz_rows=None,
+ sort=True):
+ if (left_matrix.shape[0] + right_matrix.shape[0]) > \
+ OverflowThreshold:
+ raise OverflowError
+ return real_build_matches(left_matrix, right_matrix, nnz_rows, sort)
+ return wrapper
+
+ def do_test_with(OverflowThreshold):
+ nonlocal sg # allows reference to sg, as sg will be modified below
+ # Now let us mock sg._build_matches:
+ sg._build_matches = Mock(side_effect=mock_build_matches(OverflowThreshold))
+ sg.clear_data()
+ matches_auto = fix_row_order(sg.match_strings(df1, n_blocks=None))
+ pd.testing.assert_series_equal(sg.master, df1)
+ pd.testing.assert_frame_equal(matches, matches_auto)
+ self.assertEqual(sg._config.n_blocks, None)
+ # Note that _build_matches is called more than once if and only if
+ # a split occurred (that is, there was more than one pair of
+ # matrix-blocks multiplied)
+ if len(sg._left_Series) + len(sg._right_Series) > \
+ OverflowThreshold:
+ # Assert that split occurred:
+ self.assertGreater(sg._build_matches.call_count, 1)
+ else:
+ # Assert that split did not occur:
+ self.assertEqual(sg._build_matches.call_count, 1)
+
+ # now test auto blocking by forcing an OverflowError when the
+ # combined Series' lengths is greater than 10, 5, 3, 2
+
+ do_test_with(OverflowThreshold=100) # does not trigger auto blocking
+ do_test_with(OverflowThreshold=10)
+ do_test_with(OverflowThreshold=5)
+ do_test_with(OverflowThreshold=3)
+ do_test_with(OverflowThreshold=2)
+
+ def test_n_blocks_single_DataFrame(self):
+ """tests whether manual blocking yields consistent results"""
+ sort_cols = ['right_index', 'left_index']
+
+ def fix_row_order(df):
+ return df.sort_values(sort_cols).reset_index(drop=True)
+
+ simple_example = SimpleExample()
+ df1 = simple_example.customers_df2['Customer Name']
+
+ matches11 = fix_row_order(match_strings(df1, min_similarity=0.1))
+
+ matches12 = fix_row_order(
+ match_strings(df1, n_blocks=(1, 2), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches12)
+
+ matches13 = fix_row_order(
+ match_strings(df1, n_blocks=(1, 3), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches13)
+
+ matches14 = fix_row_order(
+ match_strings(df1, n_blocks=(1, 4), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches14)
+
+ matches15 = fix_row_order(
+ match_strings(df1, n_blocks=(1, 5), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches15)
+
+ matches16 = fix_row_order(
+ match_strings(df1, n_blocks=(1, 6), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches16)
+
+ matches17 = fix_row_order(
+ match_strings(df1, n_blocks=(1, 7), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches17)
+
+ matches18 = fix_row_order(
+ match_strings(df1, n_blocks=(1, 8), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches18)
+
+ matches21 = fix_row_order(
+ match_strings(df1, n_blocks=(2, 1), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches21)
+
+ matches22 = fix_row_order(
+ match_strings(df1, n_blocks=(2, 2), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches22)
+
+ matches32 = fix_row_order(
+ match_strings(df1, n_blocks=(3, 2), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches32)
+
+ # Create a custom wrapper for this StringGrouper instance's
+ # _build_matches() method which will later be used to
+ # mock _build_matches().
+ # Note that we have to define the wrapper here because
+ # _build_matches() is a non-static function of StringGrouper
+ # and needs access to the specific StringGrouper instance sg
+ # created here.
+ sg = StringGrouper(df1, min_similarity=0.1)
+
+ def mock_build_matches(OverflowThreshold,
+ real_build_matches=sg._build_matches):
+ def wrapper(left_matrix,
+ right_matrix,
+ nnz_rows=None,
+ sort=True):
+ if (left_matrix.shape[0] + right_matrix.shape[0]) > \
+ OverflowThreshold:
+ raise OverflowError
+ return real_build_matches(left_matrix, right_matrix, nnz_rows, sort)
+ return wrapper
+
+ def test_overflow_error_with(OverflowThreshold, n_blocks):
+ nonlocal sg
+ sg._build_matches = Mock(side_effect=mock_build_matches(OverflowThreshold))
+ sg.clear_data()
+ max_left_block_size = (len(df1)//n_blocks[0]
+ + (1 if len(df1) % n_blocks[0] > 0 else 0))
+ max_right_block_size = (len(df1)//n_blocks[1]
+ + (1 if len(df1) % n_blocks[1] > 0 else 0))
+ if (max_left_block_size + max_right_block_size) > OverflowThreshold:
+ with self.assertRaises(Exception):
+ _ = sg.match_strings(df1, n_blocks=n_blocks)
+ else:
+ matches_manual = fix_row_order(sg.match_strings(df1, n_blocks=n_blocks))
+ pd.testing.assert_frame_equal(matches11, matches_manual)
+
+ test_overflow_error_with(OverflowThreshold=100, n_blocks=(1, 1))
+ test_overflow_error_with(OverflowThreshold=10, n_blocks=(1, 1))
+ test_overflow_error_with(OverflowThreshold=10, n_blocks=(2, 1))
+ test_overflow_error_with(OverflowThreshold=10, n_blocks=(1, 2))
+ test_overflow_error_with(OverflowThreshold=10, n_blocks=(4, 4))
+
+ def test_n_blocks_both_DataFrames(self):
+ """tests whether manual blocking yields consistent results"""
+ sort_cols = ['right_index', 'left_index']
+
+ def fix_row_order(df):
+ return df.sort_values(sort_cols).reset_index(drop=True)
+
+ simple_example = SimpleExample()
+ df1 = simple_example.customers_df['Customer Name']
+ df2 = simple_example.customers_df2['Customer Name']
+
+ matches11 = fix_row_order(match_strings(df1, df2, min_similarity=0.1))
+
+ matches12 = fix_row_order(
+ match_strings(df1, df2, n_blocks=(1, 2), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches12)
+
+ matches13 = fix_row_order(
+ match_strings(df1, df2, n_blocks=(1, 3), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches13)
+
+ matches14 = fix_row_order(
+ match_strings(df1, df2, n_blocks=(1, 4), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches14)
+
+ matches15 = fix_row_order(
+ match_strings(df1, df2, n_blocks=(1, 5), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches15)
+
+ matches16 = fix_row_order(
+ match_strings(df1, df2, n_blocks=(1, 6), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches16)
+
+ matches17 = fix_row_order(
+ match_strings(df1, df2, n_blocks=(1, 7), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches17)
+
+ matches18 = fix_row_order(
+ match_strings(df1, df2, n_blocks=(1, 8), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches18)
+
+ matches21 = fix_row_order(
+ match_strings(df1, df2, n_blocks=(2, 1), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches21)
+
+ matches22 = fix_row_order(
+ match_strings(df1, df2, n_blocks=(2, 2), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches22)
+
+ matches32 = fix_row_order(
+ match_strings(df1, df2, n_blocks=(3, 2), min_similarity=0.1))
+ pd.testing.assert_frame_equal(matches11, matches32)
+
+ def test_n_blocks_bad_option_value(self):
+ """Tests that bad option values for n_blocks are caught"""
+ simple_example = SimpleExample()
+ df1 = simple_example.customers_df2['Customer Name']
+ with self.assertRaises(Exception):
+ _ = match_strings(df1, n_blocks=2)
+ with self.assertRaises(Exception):
+ _ = match_strings(df1, n_blocks=(0, 2))
+ with self.assertRaises(Exception):
+ _ = match_strings(df1, n_blocks=(1, 2.5))
+ with self.assertRaises(Exception):
+ _ = match_strings(df1, n_blocks=(1, 2, 3))
+ with self.assertRaises(Exception):
+ _ = match_strings(df1, n_blocks=(1, ))
+
+ def test_tfidf_dtype_bad_option_value(self):
+ """Tests that bad option values for n_blocks are caught"""
+ simple_example = SimpleExample()
+ df1 = simple_example.customers_df2['Customer Name']
+ with self.assertRaises(Exception):
+ _ = match_strings(df1, tfidf_matrix_dtype=None)
+ with self.assertRaises(Exception):
+ _ = match_strings(df1, tfidf_matrix_dtype=0)
+ with self.assertRaises(Exception):
+ _ = match_strings(df1, tfidf_matrix_dtype='whatever')
+
def test_compute_pairwise_similarities(self):
"""tests the high-level function compute_pairwise_similarities"""
simple_example = SimpleExample()
@@ -131,6 +378,10 @@ def test_compute_pairwise_similarities(self):
],
name='similarity'
)
+ expected_result = expected_result.astype(np.float32)
+ pd.testing.assert_series_equal(expected_result, similarities)
+ sg = StringGrouper(df1, df2)
+ similarities = sg.compute_pairwise_similarities(df1, df2)
pd.testing.assert_series_equal(expected_result, similarities)
def test_compute_pairwise_similarities_data_integrity(self):
@@ -197,14 +448,17 @@ def test_match_strings(self, mock_StringGouper):
mock_StringGrouper_instance.get_matches.assert_called_once()
self.assertEqual(df, 'whatever')
- @patch('string_grouper.string_grouper.StringGrouper._symmetrize_matches_list')
- def test_match_list_symmetry_without_symmetrize_function(self, mock_symmetrize_matches_list):
- """mocks StringGrouper._symmetrize_matches_list so that this test fails whenever _matches_list is
+ @patch(
+ 'string_grouper.string_grouper.StringGrouper._symmetrize_matrix',
+ side_effect=mock_symmetrize_matrix
+ )
+ def test_match_list_symmetry_without_symmetrize_function(self, mock_symmetrize_matrix_param):
+ """mocks StringGrouper._symmetrize_matches_list so that this test fails whenever _matches_list is
**partially** symmetric which often occurs when the kwarg max_n_matches is too small"""
simple_example = SimpleExample()
df = simple_example.customers_df2['Customer Name']
sg = StringGrouper(df, max_n_matches=2).fit()
- mock_symmetrize_matches_list.assert_called_once()
+ mock_symmetrize_matrix_param.assert_called_once()
# obtain the upper and lower triangular parts of the matrix of matches:
upper = sg._matches_list[sg._matches_list['master_side'] < sg._matches_list['dupe_side']]
lower = sg._matches_list[sg._matches_list['master_side'] > sg._matches_list['dupe_side']]
@@ -213,7 +467,7 @@ def test_match_list_symmetry_without_symmetrize_function(self, mock_symmetrize_m
# obtain the intersection between upper and upper_prime:
intersection = upper_prime.merge(upper, how='inner', on=['master_side', 'dupe_side'])
# if the intersection is empty then _matches_list is completely non-symmetric (this is acceptable)
- # if the intersection is not empty then at least some matches are repeated.
+ # if the intersection is not empty then at least some matches are repeated.
# To make sure all (and not just some) matches are repeated, the lengths of
# upper, upper_prime and their intersection should be identical.
self.assertFalse(intersection.empty or len(upper) == len(upper_prime) == len(intersection))
@@ -231,38 +485,53 @@ def test_match_list_symmetry_with_symmetrize_function(self):
# Obtain the intersection between upper and upper_prime:
intersection = upper_prime.merge(upper, how='inner', on=['master_side', 'dupe_side'])
# If the intersection is empty this means _matches_list is completely non-symmetric (this is acceptable)
- # If the intersection is not empty this means at least some matches are repeated.
+ # If the intersection is not empty this means at least some matches are repeated.
# To make sure all (and not just some) matches are repeated, the lengths of
# upper, upper_prime and their intersection should be identical.
self.assertTrue(intersection.empty or len(upper) == len(upper_prime) == len(intersection))
- def test_match_list_diagonal(self):
+ @patch(
+ 'string_grouper.string_grouper.StringGrouper._fix_diagonal',
+ side_effect=mock_symmetrize_matrix
+ )
+ def test_match_list_diagonal_without_the_fix(self, mock_fix_diagonal):
"""test fails whenever _matches_list's number of self-joins is not equal to the number of strings"""
# This bug is difficult to reproduce -- I mostly encounter it while working with very large datasets;
# for small datasets setting max_n_matches=1 reproduces the bug
simple_example = SimpleExample()
df = simple_example.customers_df['Customer Name']
matches = match_strings(df, max_n_matches=1)
+ mock_fix_diagonal.assert_called_once()
num_self_joins = len(matches[matches['left_index'] == matches['right_index']])
num_strings = len(df)
self.assertNotEqual(num_self_joins, num_strings)
+ def test_match_list_diagonal(self):
+ """This test ensures that all self-joins are present"""
+ # This bug is difficult to reproduce -- I mostly encounter it while working with very large datasets;
+ # for small datasets setting max_n_matches=1 reproduces the bug
+ simple_example = SimpleExample()
+ df = simple_example.customers_df['Customer Name']
+ matches = match_strings(df, max_n_matches=1)
+ num_self_joins = len(matches[matches['left_index'] == matches['right_index']])
+ num_strings = len(df)
+ self.assertEqual(num_self_joins, num_strings)
+
def test_zero_min_similarity(self):
- """Since sparse matrices exclude zero elements, this test ensures that zero similarity matches are
+ """Since sparse matrices exclude zero elements, this test ensures that zero similarity matches are
returned when min_similarity <= 0. A bug related to this was first pointed out by @nbcvijanovic"""
simple_example = SimpleExample()
s_master = simple_example.customers_df['Customer Name']
s_dup = simple_example.whatever_series_1
- matches = match_strings(s_master, s_dup, max_n_matches=len(s_master), min_similarity=0)
+ matches = match_strings(s_master, s_dup, min_similarity=0)
pd.testing.assert_frame_equal(simple_example.expected_result_with_zeroes, matches)
def test_zero_min_similarity_small_max_n_matches(self):
- """This test ensures that a warning is issued when n_max_matches is suspected to be too small while
+ """This test ensures that a warning is issued when n_max_matches is suspected to be too small while
min_similarity <= 0 and include_zeroes is True"""
simple_example = SimpleExample()
s_master = simple_example.customers_df['Customer Name']
s_dup = simple_example.two_strings
- warnings.simplefilter('error', UserWarning)
with self.assertRaises(Exception):
_ = match_strings(s_master, s_dup, max_n_matches=1, min_similarity=0)
@@ -276,7 +545,7 @@ def test_get_non_matches_empty_case(self):
def test_n_grams_case_unchanged(self):
"""Should return all ngrams in a string with case"""
- test_series = pd.Series(pd.Series(['aa']))
+ test_series = pd.Series(pd.Series(['aaa']))
# Explicit do not ignore case
sg = StringGrouper(test_series, ignore_case=False)
expected_result = ['McD', 'cDo', 'Don', 'ona', 'nal', 'ald', 'lds']
@@ -284,7 +553,7 @@ def test_n_grams_case_unchanged(self):
def test_n_grams_ignore_case_to_lower(self):
"""Should return all case insensitive ngrams in a string"""
- test_series = pd.Series(pd.Series(['aa']))
+ test_series = pd.Series(pd.Series(['aaa']))
# Explicit ignore case
sg = StringGrouper(test_series, ignore_case=True)
expected_result = ['mcd', 'cdo', 'don', 'ona', 'nal', 'ald', 'lds']
@@ -292,7 +561,7 @@ def test_n_grams_ignore_case_to_lower(self):
def test_n_grams_ignore_case_to_lower_with_defaults(self):
"""Should return all case insensitive ngrams in a string"""
- test_series = pd.Series(pd.Series(['aa']))
+ test_series = pd.Series(pd.Series(['aaa']))
# Implicit default case (i.e. default behaviour)
sg = StringGrouper(test_series)
expected_result = ['mcd', 'cdo', 'don', 'ona', 'nal', 'ald', 'lds']
@@ -302,7 +571,7 @@ def test_build_matrix(self):
"""Should create a csr matrix only master"""
test_series = pd.Series(['foo', 'bar', 'baz'])
sg = StringGrouper(test_series)
- master, dupe = sg._get_tf_idf_matrices()
+ master, dupe = sg._get_right_tf_idf_matrix(), sg._get_left_tf_idf_matrix()
c = csr_matrix([[0., 0., 1.],
[1., 0., 0.],
[0., 1., 0.]])
@@ -314,7 +583,7 @@ def test_build_matrix_master_and_duplicates(self):
test_series_1 = pd.Series(['foo', 'bar', 'baz'])
test_series_2 = pd.Series(['foo', 'bar', 'bop'])
sg = StringGrouper(test_series_1, test_series_2)
- master, dupe = sg._get_tf_idf_matrices()
+ master, dupe = sg._get_right_tf_idf_matrix(), sg._get_left_tf_idf_matrix()
master_expected = csr_matrix([[0., 0., 0., 1.],
[1., 0., 0., 0.],
[0., 1., 0., 0.]])
@@ -330,12 +599,12 @@ def test_build_matches(self):
test_series_1 = pd.Series(['foo', 'bar', 'baz'])
test_series_2 = pd.Series(['foo', 'bar', 'bop'])
sg = StringGrouper(test_series_1, test_series_2)
- master, dupe = sg._get_tf_idf_matrices()
+ master, dupe = sg._get_right_tf_idf_matrix(), sg._get_left_tf_idf_matrix()
expected_matches = np.array([[1., 0., 0.],
[0., 1., 0.],
[0., 0., 0.]])
- np.testing.assert_array_equal(expected_matches, sg._build_matches(master, dupe).toarray())
+ np.testing.assert_array_equal(expected_matches, sg._build_matches(master, dupe)[0].toarray())
def test_build_matches_list(self):
"""Should create the cosine similarity matrix of two series"""
@@ -347,6 +616,7 @@ def test_build_matches_list(self):
dupe_side = [0, 1]
similarity = [1.0, 1.0]
expected_df = pd.DataFrame({'master_side': master, 'dupe_side': dupe_side, 'similarity': similarity})
+ expected_df.loc[:, 'similarity'] = expected_df.loc[:, 'similarity'].astype(sg._config.tfidf_matrix_dtype)
pd.testing.assert_frame_equal(expected_df, sg._matches_list)
def test_case_insensitive_build_matches_list(self):
@@ -359,6 +629,7 @@ def test_case_insensitive_build_matches_list(self):
dupe_side = [0, 1]
similarity = [1.0, 1.0]
expected_df = pd.DataFrame({'master_side': master, 'dupe_side': dupe_side, 'similarity': similarity})
+ expected_df.loc[:, 'similarity'] = expected_df.loc[:, 'similarity'].astype(sg._config.tfidf_matrix_dtype)
pd.testing.assert_frame_equal(expected_df, sg._matches_list)
def test_get_matches_two_dataframes(self):
@@ -373,6 +644,7 @@ def test_get_matches_two_dataframes(self):
expected_df = pd.DataFrame({'left_index': left_index, 'left_side': left_side,
'similarity': similarity,
'right_side': right_side, 'right_index': right_index})
+ expected_df.loc[:, 'similarity'] = expected_df.loc[:, 'similarity'].astype(sg._config.tfidf_matrix_dtype)
pd.testing.assert_frame_equal(expected_df, sg.get_matches())
def test_get_matches_single(self):
@@ -381,12 +653,13 @@ def test_get_matches_single(self):
sg = sg.fit()
left_side = ['foo', 'foo', 'bar', 'baz', 'foo', 'foo']
right_side = ['foo', 'foo', 'bar', 'baz', 'foo', 'foo']
- left_index = [0, 0, 1, 2, 3, 3]
- right_index = [0, 3, 1, 2, 0, 3]
+ left_index = [0, 3, 1, 2, 0, 3]
+ right_index = [0, 0, 1, 2, 3, 3]
similarity = [1.0, 1.0, 1.0, 1.0, 1.0, 1.0]
expected_df = pd.DataFrame({'left_index': left_index, 'left_side': left_side,
'similarity': similarity,
'right_side': right_side, 'right_index': right_index})
+ expected_df.loc[:, 'similarity'] = expected_df.loc[:, 'similarity'].astype(sg._config.tfidf_matrix_dtype)
pd.testing.assert_frame_equal(expected_df, sg.get_matches())
def test_get_matches_1_series_1_id_series(self):
@@ -395,15 +668,16 @@ def test_get_matches_1_series_1_id_series(self):
sg = StringGrouper(test_series_1, master_id=test_series_id_1)
sg = sg.fit()
left_side = ['foo', 'foo', 'bar', 'baz', 'foo', 'foo']
- left_side_id = ['A0', 'A0', 'A1', 'A2', 'A3', 'A3']
- left_index = [0, 0, 1, 2, 3, 3]
+ left_side_id = ['A0', 'A3', 'A1', 'A2', 'A0', 'A3']
+ left_index = [0, 3, 1, 2, 0, 3]
right_side = ['foo', 'foo', 'bar', 'baz', 'foo', 'foo']
- right_side_id = ['A0', 'A3', 'A1', 'A2', 'A0', 'A3']
- right_index = [0, 3, 1, 2, 0, 3]
+ right_side_id = ['A0', 'A0', 'A1', 'A2', 'A3', 'A3']
+ right_index = [0, 0, 1, 2, 3, 3]
similarity = [1.0, 1.0, 1.0, 1.0, 1.0, 1.0]
expected_df = pd.DataFrame({'left_index': left_index, 'left_side': left_side, 'left_id': left_side_id,
'similarity': similarity,
'right_id': right_side_id, 'right_side': right_side, 'right_index': right_index})
+ expected_df.loc[:, 'similarity'] = expected_df.loc[:, 'similarity'].astype(sg._config.tfidf_matrix_dtype)
pd.testing.assert_frame_equal(expected_df, sg.get_matches())
def test_get_matches_2_series_2_id_series(self):
@@ -423,6 +697,7 @@ def test_get_matches_2_series_2_id_series(self):
expected_df = pd.DataFrame({'left_index': left_index, 'left_side': left_side, 'left_id': left_side_id,
'similarity': similarity,
'right_id': right_side_id, 'right_side': right_side, 'right_index': right_index})
+ expected_df.loc[:, 'similarity'] = expected_df.loc[:, 'similarity'].astype(sg._config.tfidf_matrix_dtype)
pd.testing.assert_frame_equal(expected_df, sg.get_matches())
def test_get_matches_raises_exception_if_unexpected_options_given(self):
@@ -469,6 +744,61 @@ def test_get_groups_single_df_group_rep_default(self):
ignore_index=True
)
)
+ sg = StringGrouper(customers_df['Customer Name'])
+ pd.testing.assert_series_equal(
+ simple_example.expected_result_centroid,
+ sg.group_similar_strings(
+ customers_df['Customer Name'],
+ min_similarity=0.6,
+ ignore_index=True
+ )
+ )
+
+ def test_get_groups_single_valued_series(self):
+ """This test ensures that get_groups() returns a single-valued DataFrame or Series object
+ since the input-series is also single-valued. This test was created in response to a bug discovered
+ by George Walker"""
+ pd.testing.assert_frame_equal(
+ pd.DataFrame([(0, "hello")], columns=['group_rep_index', 'group_rep']),
+ group_similar_strings(
+ pd.Series(["hello"]),
+ min_similarity=0.6
+ )
+ )
+ pd.testing.assert_series_equal(
+ pd.Series(["hello"], name='group_rep'),
+ group_similar_strings(
+ pd.Series(["hello"]),
+ min_similarity=0.6,
+ ignore_index=True
+ )
+ )
+ pd.testing.assert_frame_equal(
+ pd.DataFrame([(0, "hello")], columns=['most_similar_index', 'most_similar_master']),
+ match_most_similar(
+ pd.Series(["hello"]),
+ pd.Series(["hello"]),
+ min_similarity=0.6
+ )
+ )
+ pd.testing.assert_frame_equal(
+ pd.DataFrame([(0, "hello")], columns=['most_similar_index', 'most_similar_master']),
+ match_most_similar(
+ pd.Series(["hello"]),
+ pd.Series(["hello"]),
+ min_similarity=0.6,
+ max_n_matches=20
+ )
+ )
+ pd.testing.assert_series_equal(
+ pd.Series(["hello"], name='most_similar_master'),
+ match_most_similar(
+ pd.Series(["hello"]),
+ pd.Series(["hello"]),
+ min_similarity=0.6,
+ ignore_index=True
+ )
+ )
def test_get_groups_single_df_keep_index(self):
"""Should return a pd.Series object with the same length as the original df. The series object will contain
@@ -542,6 +872,8 @@ def test_get_groups_two_df(self):
result = sg.get_groups()
expected_result = pd.Series(['foooo', 'bar', 'baz', 'foooo'], name='most_similar_master')
pd.testing.assert_series_equal(expected_result, result)
+ result = sg.match_most_similar(test_series_1, test_series_2, max_n_matches=3)
+ pd.testing.assert_series_equal(expected_result, result)
def test_get_groups_2_string_series_2_id_series(self):
"""Should return a pd.DataFrame object with the length of the dupes. The series will contain the master string
@@ -610,9 +942,9 @@ def test_get_groups_4_df_same_similarity(self):
test_series_2 = pd.Series(['foooo', 'bar', 'baz', 'foooob'])
test_series_id_1 = pd.Series(['A0', 'A1', 'A2', 'A3'])
test_series_id_2 = pd.Series(['B0', 'B1', 'B2', 'B3'])
- sg = StringGrouper(test_series_1,
- test_series_2,
- master_id=test_series_id_1,
+ sg = StringGrouper(test_series_1,
+ test_series_2,
+ master_id=test_series_id_1,
duplicates_id=test_series_id_2,
ignore_index=True)
sg = sg.fit()
diff --git a/string_grouper_utils/string_grouper_utils.py b/string_grouper_utils/string_grouper_utils.py
index 11803a32..e674367b 100644
--- a/string_grouper_utils/string_grouper_utils.py
+++ b/string_grouper_utils/string_grouper_utils.py
@@ -1,7 +1,7 @@
-import numpy as np
import pandas as pd
from typing import List, Optional, Union
from dateutil.parser import parse
+from dateutil.tz import UTC
from numbers import Number
from datetime import datetime
import re
@@ -137,19 +137,19 @@ def get_column(col: Union[str, int, List[Union[str, int]]], data: pd.DataFrame):
def parse_timestamps(timestamps: pd.Series, parserinfo=None, **kwargs) -> pd.Series:
- error_msg = f"timestamps must be a Series of date-like or datetime-like strings"
- error_msg += f" or datetime datatype or pandas Timestamp datatype or numbers"
+ error_msg = "timestamps must be a Series of date-like or datetime-like strings"
+ error_msg += " or datetime datatype or pandas Timestamp datatype or numbers"
if is_series_of_type(str, timestamps):
# if any of the strings is not datetime-like raise an exception
if timestamps.to_frame().applymap(is_date).squeeze().all():
# convert strings to numpy datetime64
- return timestamps.transform(lambda x: np.datetime64(parse(x, parserinfo, **kwargs)))
+ return timestamps.transform(lambda x: parse(x, parserinfo, **kwargs).astimezone(UTC))
elif is_series_of_type(type(pd.Timestamp('15-1-2000')), timestamps):
# convert pandas Timestamps to numpy datetime64
return timestamps.transform(lambda x: x.to_numpy())
elif is_series_of_type(datetime, timestamps):
# convert python datetimes to numpy datetime64
- return timestamps.transform(lambda x: np.datetime64(x))
+ return timestamps.transform(lambda x: x.astimezone(UTC))
elif is_series_of_type(Number, timestamps):
return timestamps
raise Exception(error_msg)
diff --git a/string_grouper_utils/test/test_string_grouper_utils.py b/string_grouper_utils/test/test_string_grouper_utils.py
index 3798e3cd..0c8a8ee4 100644
--- a/string_grouper_utils/test/test_string_grouper_utils.py
+++ b/string_grouper_utils/test/test_string_grouper_utils.py
@@ -1,8 +1,8 @@
import unittest
import pandas as pd
from dateutil.parser import parse
-from string_grouper_utils.string_grouper_utils import new_group_rep_by_earliest_timestamp, new_group_rep_by_completeness, \
- new_group_rep_by_highest_weight
+from string_grouper_utils.string_grouper_utils import new_group_rep_by_earliest_timestamp, \
+ new_group_rep_by_completeness, new_group_rep_by_highest_weight
class SimpleExample(object):
diff --git a/time_match_strings.py b/time_match_strings.py
new file mode 100644
index 00000000..ee87b204
--- /dev/null
+++ b/time_match_strings.py
@@ -0,0 +1,63 @@
+import pandas as pd
+import numpy as np
+from string_grouper import match_strings
+import random
+import time
+import os
+
+# mem_limit = '1G'
+# procgov = r'C:\Users\heamu\Source\Repos\process-governor\ProcessGovernor\bin\x64\Debug\procgov.exe'
+# os.popen(f'{procgov} -r -m {mem_limit} -p {os.getpid()}')
+# time.sleep(1)
+progress = 0
+do_print = True
+companies = pd.read_csv('data/sec__edgar_company_info.csv')
+x0 = 10000
+Nx = 10000
+dNx = 1000
+Nx2 = 500000
+dNx2 = 50000
+y0 = 10000
+Ny = 10000
+dNy = 10000
+ns = 10
+# X = np.append(np.arange(dNx, Nx + 1, dNx), np.arange(dNx2 + dNx2, Nx2 + 1, dNx2))
+X = np.arange(x0, Nx + 1, dNx)
+Y = np.arange(y0, Ny + 1, dNy)
+means = np.full((len(X), len(Y)), 0)
+for s in range(ns):
+ dgrid = []
+ i = 1
+ _ = print('[', flush=True, end='') if do_print else None
+ for x in X:
+ left_df = companies['Company Name'].iloc[random.sample(range(len(companies)), k = x)]
+ if i > 1:
+ _ = print(', ', flush=True) if do_print else None
+ dseries = []
+ stdseries = []
+ _ = print('[', flush=True, end='') if do_print else None
+ j = 1
+ for y in Y:
+ if j > 1:
+ _ = print(', ', flush=True, end='') if do_print else None
+ right_df = companies['Company Name'].iloc[random.sample(range(len(companies)), k = y)]
+ t0 = time.time()
+ _ = match_strings(right_df, left_df, n_blocks=(1, 1))
+ t1 = time.time()
+ dseries += [(t1 - t0)/60]
+ progress += 1.0/(ns*len(X)*len(Y))
+ # print(f'Progress {progress:.1%}', end='\x1b[1K\r')
+ _ = print(f'{dseries[-1]}', flush=True, end='') if do_print else None
+ # _ = print('.', flush=True, end='') if not do_print else None
+ j += 1
+ _ = print(']', flush=True, end='') if do_print else None
+ dgrid += [dseries]
+ i += 1
+ # _ = print(f'{i}/{len(X)}', flush=True) if not do_print else None
+ _ = print(']', flush=True) if do_print else None
+ means = (np.asarray(dgrid) + s*means)/(s + 1)
+ with open(f'runtime_means_x_{x0}-{Nx}_y_{y0}-{Ny}.npy', 'wb') as f:
+ np.save(f, means)
+ np.save(f, X)
+ np.save(f, Y)
+ #send_me_mail()