|
74 | 74 | "\n", |
75 | 75 | "Tidy data aren't going to be appropriate *every* time and in every case, but they're a really, really good default for tabular data. Once you use it as your default, it's easier to think about how to perform subsequent operations.\n", |
76 | 76 | "\n", |
77 | | - "Having said that tidy data are great, they are, but one of **pandas**' advantages relative to other data analysis libraries is that it isn't *too* tied to tidy data and can navigate awkward non-tidy data manipulation tasks happily too." |
| 77 | + "Having said that tidy data are great, they are, but one of **pandas**' advantages relative to other data analysis libraries is that it isn't *too* tied to tidy data and can navigate awkward non-tidy data manipulation tasks happily too.\n", |
| 78 | + "\n", |
| 79 | + "There are two common problems you find in data that are ingested that make them not tidy:\n", |
| 80 | + "\n", |
| 81 | + "1. A variable might be spread across multiple columns.\n", |
| 82 | + "2. An observation might be scattered across multiple rows.\n", |
| 83 | + "\n", |
| 84 | + "For the former, we need to \"melt\" the wide data, with multiple columns, into long data.\n", |
| 85 | + "\n", |
| 86 | + "For the latter, we need to unstack or pivot the multiple rows into columns (ie go from long to wide.)\n", |
| 87 | + "\n", |
| 88 | + "We'll see both below." |
78 | 89 | ] |
79 | 90 | }, |
80 | 91 | { |
81 | 92 | "cell_type": "markdown", |
82 | 93 | "id": "deb8cf13", |
83 | 94 | "metadata": {}, |
84 | 95 | "source": [ |
85 | | - "## Make Data Tidy with **pandas**" |
| 96 | + "## Tools to Make Data Tidy with **pandas**" |
86 | 97 | ] |
87 | 98 | }, |
88 | 99 | { |
|
92 | 103 | "source": [ |
93 | 104 | "### Melt\n", |
94 | 105 | "\n", |
95 | | - "`melt()` can help you go from untidy to tidy data (from wide data to long data), and is a *really* good one to remember.\n", |
| 106 | + "`melt()` can help you go from \"wider\" data to \"longer\" data, and is a *really* good one to remember.\n", |
96 | 107 | "\n", |
97 | 108 | "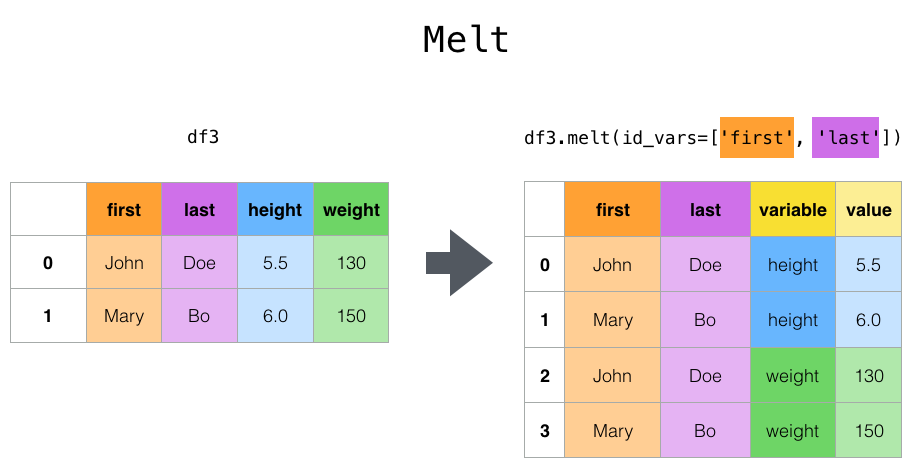\n", |
98 | 109 | "\n", |
|
132 | 143 | "Perform a `melt()` that uses `job` as the id instead of `first` and `last`.\n", |
133 | 144 | "```\n", |
134 | 145 | "\n", |
135 | | - "### Wide to Long\n", |
| 146 | + "How does this relate to tidy data? Sometimes you'll have a variable spread over multiple columns that you want to turn tidy. Let's look at this example that uses cases of [tuburculosis from the World Health Organisation](https://www.who.int/teams/global-tuberculosis-programme/data).\n", |
| 147 | + "\n", |
| 148 | + "First let's open the data and look at the top of the file." |
| 149 | + ] |
| 150 | + }, |
| 151 | + { |
| 152 | + "cell_type": "code", |
| 153 | + "execution_count": null, |
| 154 | + "id": "bfa121cf", |
| 155 | + "metadata": {}, |
| 156 | + "outputs": [], |
| 157 | + "source": [ |
| 158 | + "from pathlib import Path\n", |
| 159 | + "\n", |
| 160 | + "df_tb = pd.read_parquet(Path(\"data/who_tb_cases.parquet\"))\n", |
| 161 | + "df_tb.head()" |
| 162 | + ] |
| 163 | + }, |
| 164 | + { |
| 165 | + "cell_type": "markdown", |
| 166 | + "id": "583e419d", |
| 167 | + "metadata": {}, |
| 168 | + "source": [ |
| 169 | + "You can see that we have two columns for a single variable, year. Let's now melt this." |
| 170 | + ] |
| 171 | + }, |
| 172 | + { |
| 173 | + "cell_type": "code", |
| 174 | + "execution_count": null, |
| 175 | + "id": "dc03ccd9", |
| 176 | + "metadata": {}, |
| 177 | + "outputs": [], |
| 178 | + "source": [ |
| 179 | + "df_tb.melt(\n", |
| 180 | + " id_vars=[\"country\"],\n", |
| 181 | + " var_name=\"year\",\n", |
| 182 | + " value_vars=[\"1999\", \"2000\"],\n", |
| 183 | + " value_name=\"cases\",\n", |
| 184 | + ")" |
| 185 | + ] |
| 186 | + }, |
| 187 | + { |
| 188 | + "cell_type": "markdown", |
| 189 | + "id": "74d81b30", |
| 190 | + "metadata": {}, |
| 191 | + "source": [ |
| 192 | + "We now have one observation per row, and one variable per column: tidy!" |
| 193 | + ] |
| 194 | + }, |
| 195 | + { |
| 196 | + "cell_type": "markdown", |
| 197 | + "id": "488f5f10", |
| 198 | + "metadata": {}, |
| 199 | + "source": [ |
| 200 | + "### A simpler wide to long\n", |
136 | 201 | "\n", |
137 | 202 | "If you don't want the headscratching of `melt()`, there's also `wide_to_long()`, which is really useful for typical data cleaning cases where you have data like this:" |
138 | 203 | ] |
|
279 | 344 | "id": "39a210bb", |
280 | 345 | "metadata": {}, |
281 | 346 | "source": [ |
282 | | - "### Pivoting data from tidy to, err, untidy data\n", |
| 347 | + "### Pivoting data from long to wide\n", |
283 | 348 | "\n", |
284 | | - "At the start of this chapter, we said you should use tidy data--one row per observation, one column per variable--whenever you can. But there are times when you will want to take your lovingly prepared tidy data and pivot it into a wider format. `pivot()` and `pivot_table()` help you to do that.\n", |
| 349 | + "`pivot()` and `pivot_table()` help you to sort out data in which a single observation is scattered over multiple rows.\n", |
285 | 350 | "\n", |
286 | | - "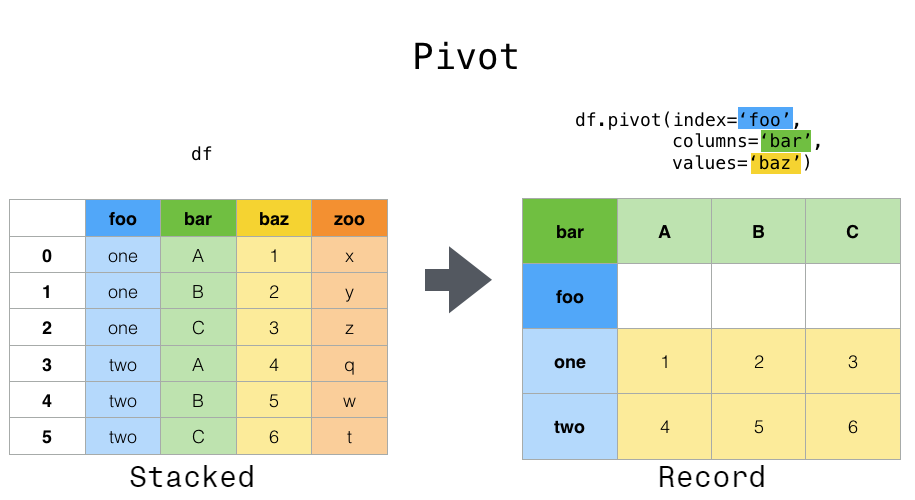\n", |
287 | | - "\n", |
288 | | - "This can be especially useful for time series data, where operations like `shift()` or `diff()` are typically applied assuming that an entry in one row follows (in time) from the one above. Here's an example:" |
| 351 | + "\n" |
| 352 | + ] |
| 353 | + }, |
| 354 | + { |
| 355 | + "cell_type": "markdown", |
| 356 | + "id": "8acbcedf", |
| 357 | + "metadata": {}, |
| 358 | + "source": [ |
| 359 | + "Here's an example dataframe where observations are spread over multiple rows:" |
289 | 360 | ] |
290 | 361 | }, |
291 | 362 | { |
292 | 363 | "cell_type": "code", |
293 | 364 | "execution_count": null, |
294 | | - "id": "405da609", |
| 365 | + "id": "fa612456", |
| 366 | + "metadata": {}, |
| 367 | + "outputs": [], |
| 368 | + "source": [ |
| 369 | + "df_tb_cp = pd.read_parquet(Path(\"data/who_tb_case_and_pop.parquet\"))\n", |
| 370 | + "df_tb_cp.head()" |
| 371 | + ] |
| 372 | + }, |
| 373 | + { |
| 374 | + "cell_type": "markdown", |
| 375 | + "id": "0d4a4077", |
| 376 | + "metadata": {}, |
| 377 | + "source": [ |
| 378 | + "You see that we have, for each year-country, \"case\" and \"population\" in different rows." |
| 379 | + ] |
| 380 | + }, |
| 381 | + { |
| 382 | + "cell_type": "markdown", |
| 383 | + "id": "e7c9ed1b", |
| 384 | + "metadata": {}, |
| 385 | + "source": [ |
| 386 | + "Now let's pivot this to see the difference:" |
| 387 | + ] |
| 388 | + }, |
| 389 | + { |
| 390 | + "cell_type": "code", |
| 391 | + "execution_count": null, |
| 392 | + "id": "e584cf37", |
| 393 | + "metadata": {}, |
| 394 | + "outputs": [], |
| 395 | + "source": [ |
| 396 | + "pivoted = df_tb_cp.pivot(\n", |
| 397 | + " index=[\"country\", \"year\"], columns=[\"type\"], values=\"count\"\n", |
| 398 | + ").reset_index()\n", |
| 399 | + "pivoted" |
| 400 | + ] |
| 401 | + }, |
| 402 | + { |
| 403 | + "cell_type": "markdown", |
| 404 | + "id": "d82307c0", |
| 405 | + "metadata": {}, |
| 406 | + "source": [ |
| 407 | + "Pivots are especially useful for time series data, where operations like `shift()` or `diff()` are typically applied assuming that an entry in one row follows (in time) from the one above. When we do `shift()` we often want to shift a single variable in time, but if a single observation (in this case a date) is over multiple rows, the timing is going go awry. Let's see an example." |
| 408 | + ] |
| 409 | + }, |
| 410 | + { |
| 411 | + "cell_type": "code", |
| 412 | + "execution_count": null, |
| 413 | + "id": "97c6d139", |
295 | 414 | "metadata": {}, |
296 | 415 | "outputs": [], |
297 | 416 | "source": [ |
|
300 | 419 | "data = {\n", |
301 | 420 | " \"value\": np.random.randn(20),\n", |
302 | 421 | " \"variable\": [\"A\"] * 10 + [\"B\"] * 10,\n", |
303 | | - " \"category\": np.random.choice([\"type1\", \"type2\", \"type3\", \"type4\"], 20),\n", |
304 | 422 | " \"date\": (\n", |
305 | | - " list(pd.date_range(\"1/1/2000\", periods=10, freq=\"M\"))\n", |
306 | | - " + list(pd.date_range(\"1/1/2000\", periods=10, freq=\"M\"))\n", |
| 423 | + " list(pd.date_range(\"1/1/2000\", periods=10, freq=\"ME\"))\n", |
| 424 | + " + list(pd.date_range(\"1/1/2000\", periods=10, freq=\"ME\"))\n", |
307 | 425 | " ),\n", |
308 | 426 | "}\n", |
309 | | - "df = pd.DataFrame(data, columns=[\"date\", \"variable\", \"category\", \"value\"])\n", |
| 427 | + "df = pd.DataFrame(data, columns=[\"date\", \"variable\", \"value\"])\n", |
310 | 428 | "df.sample(5)" |
311 | 429 | ] |
312 | 430 | }, |
|
315 | 433 | "id": "90a5b930", |
316 | 434 | "metadata": {}, |
317 | 435 | "source": [ |
318 | | - "If we just run `shift()` on this, it's going to shift variable B's and A's together even though these overlap in time. So we pivot to a wider format (and then we can shift safely)." |
| 436 | + "If we just run `shift()` on the above, it's going to shift variable B's and A's together even though these overlap in time and are different variables. So we pivot to a wider format (and then we can shift in time safely)." |
319 | 437 | ] |
320 | 438 | }, |
321 | 439 | { |
|
333 | 451 | "id": "841c4821", |
334 | 452 | "metadata": {}, |
335 | 453 | "source": [ |
336 | | - "To go back to the original structure, albeit without the `category` columns, apply `.unstack().reset_index()`.\n", |
| 454 | + "```{admonition} Exercise\n", |
| 455 | + "Why is the first entry NaN?\n", |
| 456 | + "```\n", |
| 457 | + "\n", |
337 | 458 | "\n", |
338 | 459 | "```{admonition} Exercise\n", |
339 | | - "Perform a `pivot()` that applies to both the `variable` and `category` columns. (Hint: remember that you will need to pass multiple objects via a list.)\n", |
| 460 | + "Perform a `pivot()` that applies to both the `variable` and `category` columns in the example from above where category is defined such that `df[\"category\"] = np.random.choice([\"type1\", \"type2\", \"type3\", \"type4\"], 20). (Hint: remember that you will need to pass multiple objects via a list.)\n", |
340 | 461 | "```" |
341 | 462 | ] |
342 | 463 | } |
343 | 464 | ], |
344 | 465 | "metadata": { |
345 | | - "interpreter": { |
346 | | - "hash": "9d7534ecd9fbc7d385378f8400cf4d6cb9c6175408a574f1c99c5269f08771cc" |
347 | | - }, |
348 | 466 | "jupytext": { |
349 | 467 | "cell_metadata_filter": "-all", |
350 | 468 | "encoding": "# -*- coding: utf-8 -*-", |
351 | 469 | "formats": "md:myst", |
352 | 470 | "main_language": "python" |
353 | 471 | }, |
354 | 472 | "kernelspec": { |
355 | | - "display_name": "Python 3 (ipykernel)", |
| 473 | + "display_name": ".venv", |
356 | 474 | "language": "python", |
357 | 475 | "name": "python3" |
358 | 476 | }, |
|
366 | 484 | "name": "python", |
367 | 485 | "nbconvert_exporter": "python", |
368 | 486 | "pygments_lexer": "ipython3", |
369 | | - "version": "3.10.12" |
| 487 | + "version": "3.10.0" |
370 | 488 | }, |
371 | 489 | "toc-showtags": true |
372 | 490 | }, |
|
0 commit comments