This project adds python bindings for the rust-sugiyama crate, to produce directed graph layouts similar to the GraphViz dot program.
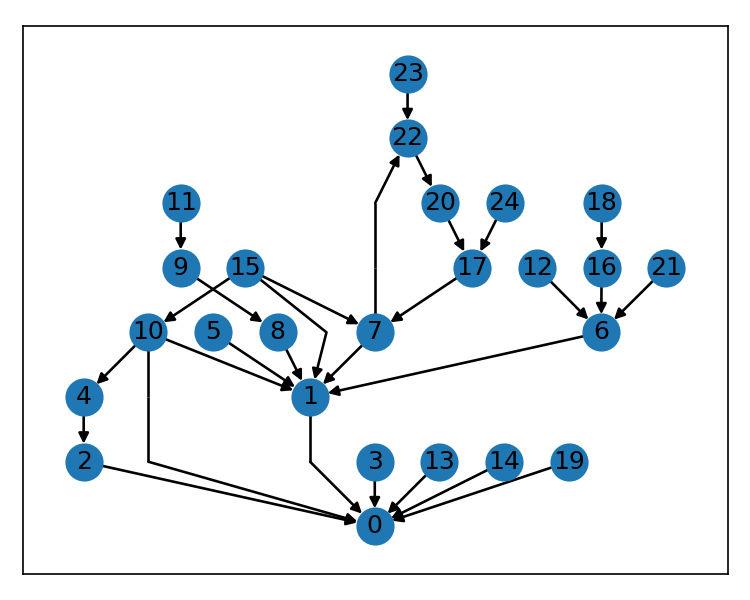
It's an implementation of Sugiyama's algorithm for displaying a layered, or hierarchical, directed graph like so:
Why?
We already have nx.nx_pydot.pydot_layout(gn, prog="dot"), grandalf.layouts.SugiyamaLayout, netgraph.get_sugiyama_layout, and rustworkx.visualization.graphviz_draw... why pile on?
All of the above either depend on either the dot program from graphviz, or the pure-python Sugiyama implementation from grandalf.
The dot program is probably the best solution to this problem that currently exists, and you should use the dot program if you are working in an environment that allows you to install graphviz and your python application only needs to render very small graphs and/or you don't need to produce them quickly.
The grandalf-based solutions are pure-python which is awesome for portability, but is slow for medium/large graphs (because python).
In addition, that project hasn't had maintenance in many years, including incorporating the 2020 Erratum from Brandes et., al on which the implementation appears to be based.
This implementation suffers from some occasions layouts where two nodes can share position, which either reveals a bug referenced in the erratum, or in the original implementation.
Anyway, the grandalf project has an excellent framework for this problem, and has a deep bench of potential mentors and prior contributors, so if you need a pure-python implementation you should support them.
This project is about having zero 3rd-party dependencies, reasonable multi-platform support (arm & x86 wheels are available for linux, windows, and mac), and speed.
In service of all three of these goals, this library is a thin wrapper around the excellent rust crate rust-sugiyama, which is likely the clearest-eyed implementation of the complex Sugiyama algorithm that's available for free on the internet.
With these shoulders to stand on, this project aims to offer very good dot-like layouts, very quickly, and with no 3rd-party runtime dependencies.
This is a great fit for folks who can't (or don't want to) install graphviz, or for web-applications that want smaller images, or for anyone needing a large directed graph layout to run fast.
import networkx as nx
from fast_sugiyama import from_edges
g = nx.gn_graph(42, seed=132, create_using=nx.DiGraph)
pos = from_edges(g.edges()).to_dict()
nx.draw_networkx(g, pos=pos, with_labels=False, node_size=150)The above example is for a networkx graph, but the fast-sugiyama package can be used with any graph library.
The return type from the to_dict() method works equally well for the rustworkx Pos2DMapping type as the networkx pos arg.
It's a {node_id: (x, y)} coordinate mapping you can take where you please.
You might also wonder why we don't emulate the networkx layout api conventions exactly (e.g., immediately return exactly one layout), and we'll talk about that a little later.
This package is pip installable.
pip install fast-sugiyamaAs with all python endeavours, you should probably be using a virtual environment for your project via conda \ mamba or venv (preferrably via uv).
uv pip install fast-sugiyamaEither of the above two install commands should work well with either ARM or x86 systems running Linux, Mac, or Windows. If you have another system and want to run this package and you can't install/build/run from the sdist or the repo please open a detailed issue and let me know what happened. Maybe I can help.
Purely for simplicity, this section will use networkx to facilitate usage demonstration and discussion.
Again, this project has no dependencies, and can be used to support high-quality layout visuals for any graph library that uses a node -> coord mapping.
The from_edges function returns a list of layouts, one for each weakly connected component of the input graph.
Each layout is a tuple containing the positions, width, height, and edges returned by the solver.
This makes it easy to deal with multi-graphs, and let's us write helper functions to support various ways of displaying separate graphs in one layout.
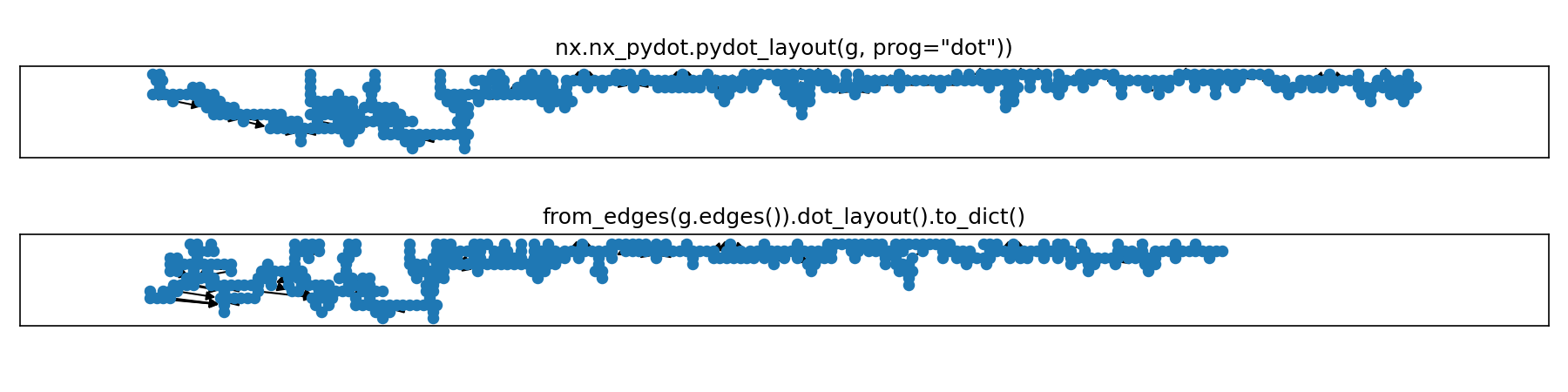
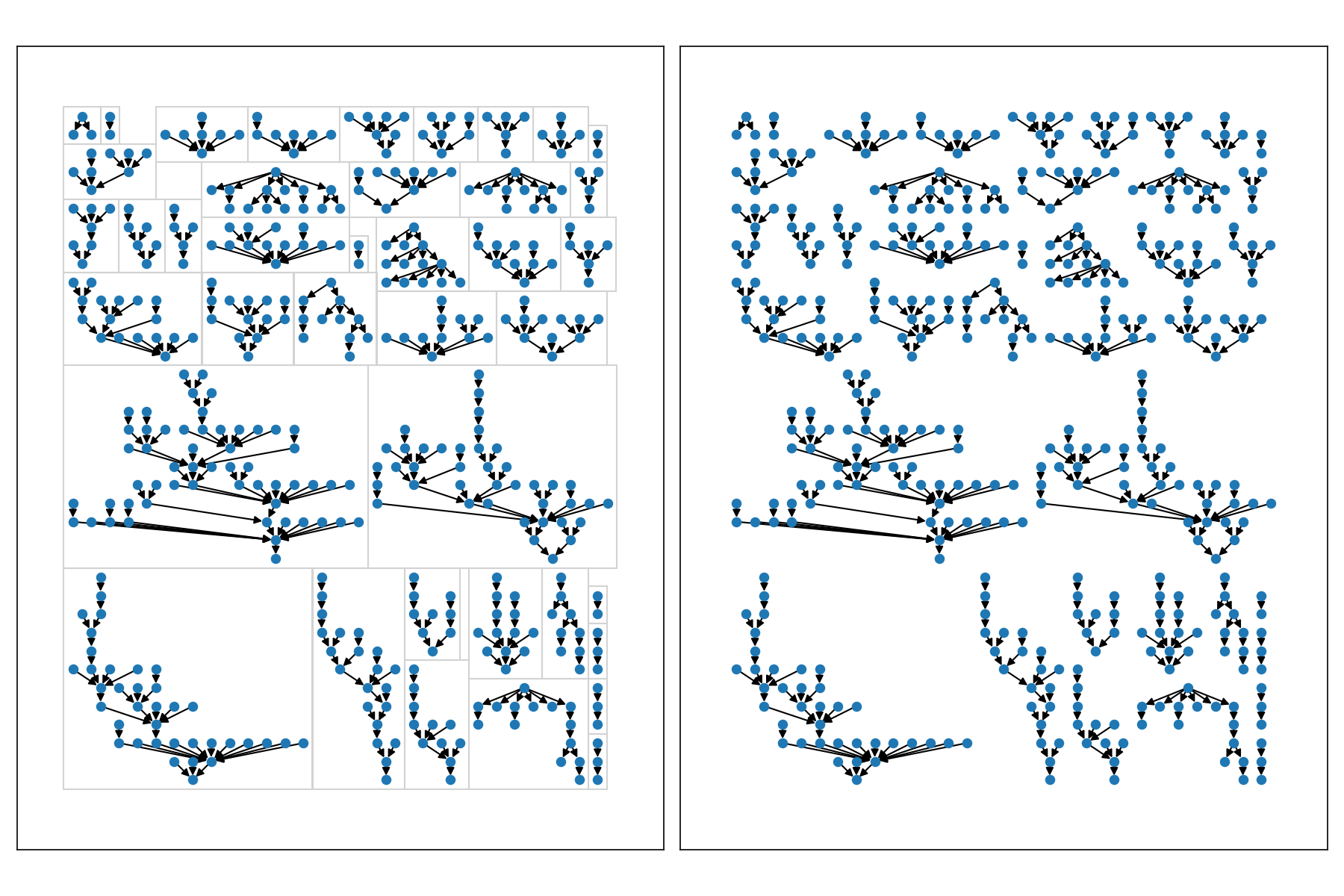
This section will use a multi-graph from the test suite that includes 40 component graphs, each with a range of sizes.
In these examples, the full graph g is made of 40 components, 435 nodes, and 395 edges.
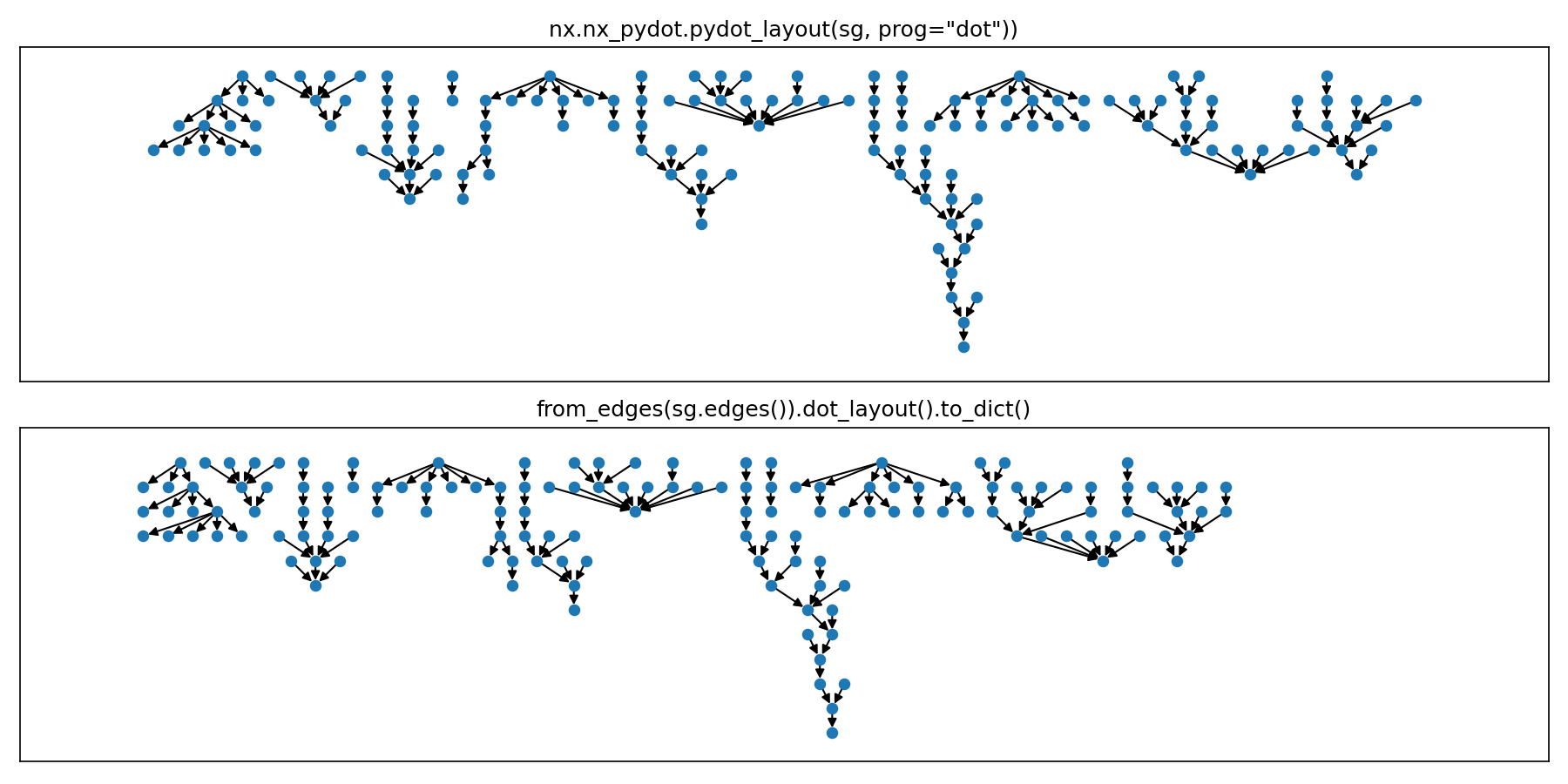
There is also a sub-set of these components called sg that includes just 12 of the 40 components.
This package has a helper function for users who would like to get a layout (or a multi-graph layout) similar to the dot program -- specifically, similar to the layout produced by nx.pydot.pydot_layout.
This layout aligns hierarchies to the top of the figure, lines them up side by side horizontally, and packs them efficiently such that nodes in neighboring graph components are separated by the same spacing interval as nodes within each component.
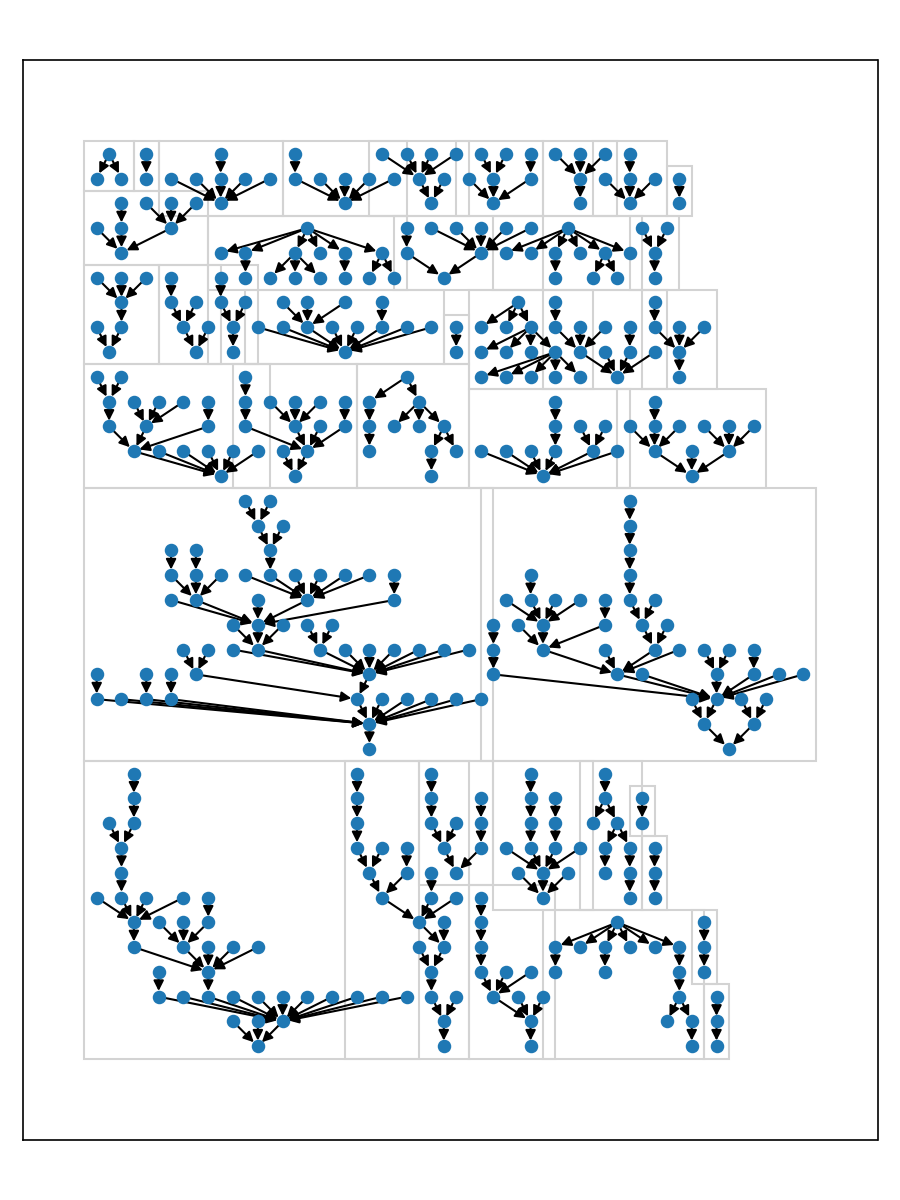
The following plot compares the layout results for a 12 component multi-graph sg.
Note that the bounding boxes of the layouts are allowed to overlap in the pydot layout, so this library supports that as well.
Lining up components horizontally quickly becomes unwieldy with the pydot layout though.
Here's all 40 components of our multi-graph g.
Users can install an optional dependency rectangle-packer to produce an improved layout that compactly packs the components.
pos = from_edges(g.edges()).rect_pack_layouts(max_width=2500).to_dict()One can even utilize the 'horizontal compact' idea from the dot-like layouts to tighten things up even further.
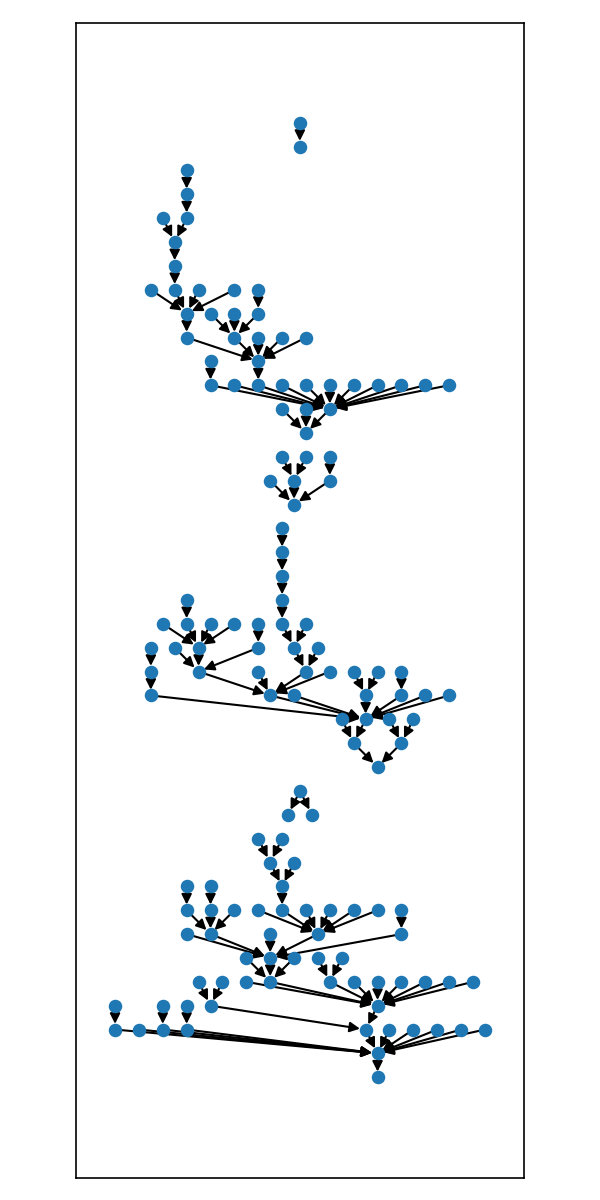
pos = from_edges(g.edges()).compact_layout(max_width=2500).to_dict()
# or equivalently
pos = (
from_edges(g.edges())
.rect_pack_layouts(max_width=2500)
.sort_horizontal()
.compact_layouts_horizontal()
)The helper layouts above simply chain a couple layout 'primitive' operations.
For example, the .dot_layout() is equivalent to .align_layouts_vertical_top().compact_layouts_horizontal()
You can chain together the built in primitives, or adapt their code into your own custom functions to obtain any arrangement.
pos = (
from_edges(g.edges())
[:6] # chaining even works after slices
.align_layouts_vertical()
.align_layouts_horizontal_center()
.to_dict()
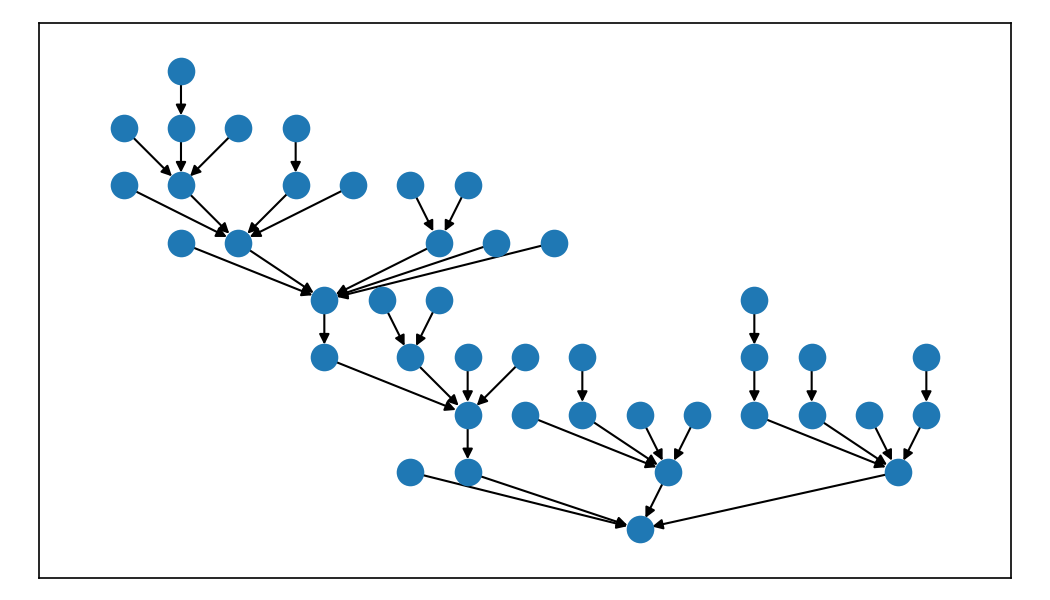
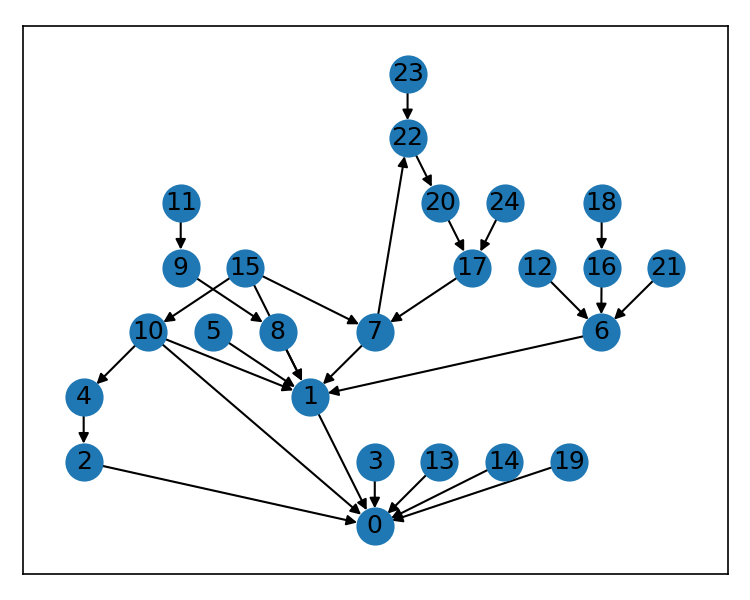
)If you're feeling wild and your graph is not simple and acyclic, you can even capture the dummy vertices and their edges from the Sugiyama solver and render those.
Here's another graph from the test suite that is directed but is not a DAG.
Let's say these edges are in a variable called edges.
g = nx.DiGraph()
g.add_edges_from(edges)
pos = from_edges(edges).to_dict()
fig, ax = plt.subplots()
nx.draw_networkx(g, pos=pos, ax=ax)But there's an issue with the edge from (15, 1) -- the algorithm created a gap for the edge to go around node 8, not behind it. We can render that edge path easily by using the edges returned by the solver in our layout.
layouts = from_edges(edges)
pos = layouts.to_dict()
nodes = list({n for edge in edges for n in edge})
g = nx.DiGraph()
for *_, edgelist in layouts: # the solver returns the edges with the dummy's included
if edgelist is not None:
g.add_edges_from(edgelist)
fig, ax = plt.subplots()
nx.draw_networkx_nodes(g, pos, ax=ax, nodelist=nodes)
nx.draw_networkx_labels(g, pos, ax=ax, labels={n: n for n in nodes})
_ = nx.draw_networkx_edges(
g,
pos,
[e for e in g.edges() if e[1] not in nodes],
arrows=False,
ax=ax,
)
_ = nx.draw_networkx_edges(
g,
pos,
[e for e in g.edges() if e[1] in nodes],
node_size=[300 if n in nodes else 0 for n in g.nodes()],
ax=ax,
)Speed is not the purpose of this package, but it is a welcome benefit of using rust for all the heavy lifting.
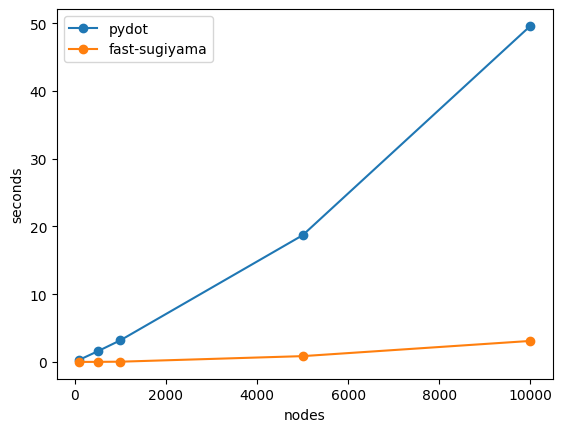
There are vanishingly few non-pathological reasons to produce a hierarchical graph plot with 5,000+ nodes, but for reasonable graph sizes, say, under 1000, this package is ~100-400x faster than using pydot.
NOTE: pydot will typically yield a nicer looking layout, and if you can install it in your environment and wait for pydot to do its serialization/deserialization, you should.
for n in [100, 500, 1000, 5000, 10000]:
g = nx.gn_graph(n, seed=42)
%timeit _ = from_edges(g.edges()).dot_layout().to_dict()
%timeit _ = nx.nx_pydot.pydot_layout(g, prog='dot')786 μs ± 6.88 μs per loop (mean ± std. dev. of 7 runs, 1,000 loops each) <-- ours @ n==100 (410x faster) 325 ms ± 9.01 ms per loop (mean ± std. dev. of 7 runs, 1 loop each) 10.9 ms ± 112 μs per loop (mean ± std. dev. of 7 runs, 100 loops each) <-- ours @ n==500 (140x faster) 1.57 s ± 27.7 ms per loop (mean ± std. dev. of 7 runs, 1 loop each) 35.8 ms ± 48.2 μs per loop (mean ± std. dev. of 7 runs, 10 loops each) <-- ours @ n==1000 (89x faster) 3.2 s ± 8.41 ms per loop (mean ± std. dev. of 7 runs, 1 loop each) 855 ms ± 5.84 ms per loop (mean ± std. dev. of 7 runs, 1 loop each) <-- ours @ n==5000 (21x faster) 18.7 s ± 234 ms per loop (mean ± std. dev. of 7 runs, 1 loop each) 3.1 s ± 23.8 ms per loop (mean ± std. dev. of 7 runs, 1 loop each) <-- ours @ n==10000 (16x faster) 49.6 s ± 1.07 s per loop (mean ± std. dev. of 7 runs, 1 loop each)
And if we plot this up...
This project contains a fork of the excellent rust-sugiyama crate, and includes minor api changes to support layouts via the python bindings.
This fork is not intended to ever be published to crates.io.
Instead anything useful we do here will be offered to the upstream crate for consideration.
Differences from the upstream project currently include:
- Compute layout width and height in layout units rather than node count
- Optionally validate the layout to prevent/detect overlapping nodes & invalid layouts
- Return dummy vertices and edges if they're part of the layout
- Remove dev-dependencies that point to personal github repos and replace with alternatives
Big thank you to the developers of the rust-sugiyama crate for doing the heavy lifting!