iCellR is an interactive R package designed to facilitate the analysis and visualization of high-throughput single-cell sequencing data. It supports a variety of single-cell technologies, including scRNA-Seq, scVDJ-Seq, scATAC-Seq, CITE-Seq, and Spatial Transcriptomics (ST).
Maintainer: Alireza Khodadadi-Jamayran
Use the latest version of iCellR (v1.6.4) for scATAC-seq and Spatial Transcriptomics (ST) analyses. Leverage the i.score function for scoring cells based on gene signatures using methods such as Tirosh, Mean, Sum, GSVA, ssgsea, Zscore, and Plage.
Explore iCellR version 1.5.5, now featuring tools for cell cycle analysis (phases G0, G1S, G2M, M, G1M, and S). See example phase, New Pseudotime Abstract KNetL (PAK map) functionality added – visualize pseudotime progression (PAK map). Perform gene-gene correlation analysis using updated visualization tools. correlations.
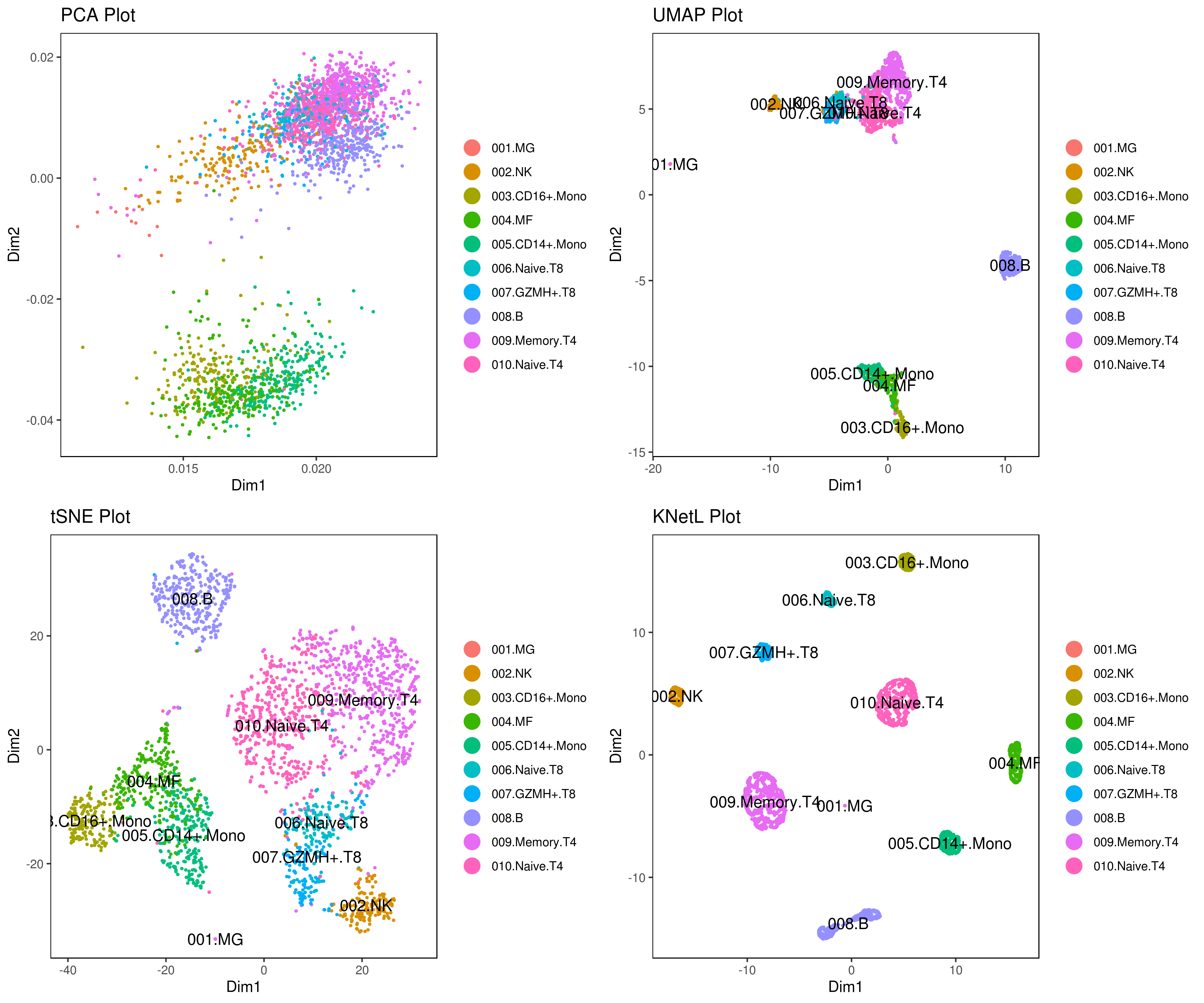
Explore the KNetL map, an advanced adjustable and dynamic dimensionality reduction method KNetL map ![]() KNetL (pronounced “nettle”) offers enhanced zooming capabilities KNetL to show significantly more detail compared to tSNE and UMAP.
KNetL (pronounced “nettle”) offers enhanced zooming capabilities KNetL to show significantly more detail compared to tSNE and UMAP.
Introducing imputation and coverage correction (CC) methods for improved gene-gene correlation analysis. (CC). Perform batch alignment using iCellR's CPCA and CCCA tools (CCCA and CPCA) methods. Expanded databases for cell type prediction now include ImmGen and MCA.
scSeqR has been renamed to iCellR, and scSeqR has been discontinued. Please use iCellR moving forward, as scSeqR is no longer supported. UMAP is added to iCellR. Interactive cell gating has been added, allowing users to select cells directly within HTML plots using Plotly.
- Link to
manualManual and Comprehensive R Archive Network (CRAN). - Gor
getting startedandtutorialsgo to our Wiki page. - Link to a video tutorial for CITE-Seq and scRNA-Seq analysis: Video
- All you need to know about KNetL map: Video
- If you are using
FlowJoorSeqGeq, they offer plugins for iCellR and other single-cell analysis tools. You can find the list of all plugins here: https://www.flowjo.com/exchange/#/ . Specifically, the iCellR plugin can be found here: https://www.flowjo.com/exchange/#/plugin/profile?id=34. Additionally, a SeqGeq Differential Expression (DE) tutorial is available to guide you through the process: SeqGeq DE tutorial
For citing iCellR use this PMID: 34353854
iCellR publications: PMID: 35660135 (scRNA-seq/KNetL) PMID: 35180378 (CITE-seq/KNetL), PMID: 34911733 (i.score and cell ranking), PMID: 34963055 (scRNA-seq), PMID 31744829 (scRNA-seq), PMID: 31934613 (bulk RNA-seq from TCGA), PMID: 32550269 (scVDJ-seq), PMID: 34135081, PMID: 33593073, PMID: 34634466, PMID: 35302059, PMID: 34353854
Single (i) Cell R package (iCellR)