-
Notifications
You must be signed in to change notification settings - Fork 317
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Add Isaac Morales blog #1606
Add Isaac Morales blog #1606
Conversation
|
Very nice, thanks! Giving @vgvassilev @LukasBreitwieser a chance to comment before merging |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Lgtm!
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Hi @imorlxs ,
Thank you very much for your work and blog. I left a few comments to strengthen the text. Let me know if you have any questions.
Best,
Lukas
| particularly in fields like cancer research, epidemiology, and social sciences. It leverages | ||
| ROOT—a framework widely used in high-energy physics—for statistical analysis, random number | ||
| generation, C++ Jupyter notebooks, and I/O operations. However, enhancing BioDynaMo’s performance | ||
| remains a key challenge. This is where this Google Summer of Code 2024 (GSoC ‘24) project comes |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Be more precise in explaining which parts of BioDynaMo need performance improvements and say why.
e.g.: the listed functionalities rely on reflection information; launching the reflection subsystem on simulation startup requires a significant amount of time and memory.
Cite this paper (https://link.springer.com/article/10.1007/s00285-024-02144-2), which describes the performance issue at startup.
Your work addresses this issue fundamentally at its root, while the paper above uses a workaround (yet effective) to improve performance.
| experiences significant runtime performance and memory usage issues. The repeated parsing of library | ||
| descriptors by Cling introduces inefficiencies that slow down the startup phase and consume excessive |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Explain the steps of cling startup and dictionary loading without C++ modules in greater detail. What exactly do you mean by "repeated parsing".
| these performance issues. C++ Modules offer an efficient on-disk representation of C++ code, | ||
| reducing the need for repeated parsing of invariant code. By implementing these modules, |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Give more details about C++ modules and how they address the challenges described before.
| The primary goal of the GSoC project was to integrate ROOT’s C++ Modules into BioDynaMo to minimize | ||
| these performance issues. C++ Modules offer an efficient on-disk representation of C++ code, | ||
| reducing the need for repeated parsing of invariant code. By implementing these modules, | ||
| the project aimed to optimize runtime memory usage and improve overall performance |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Be more precise by what you mean with "overall performance".
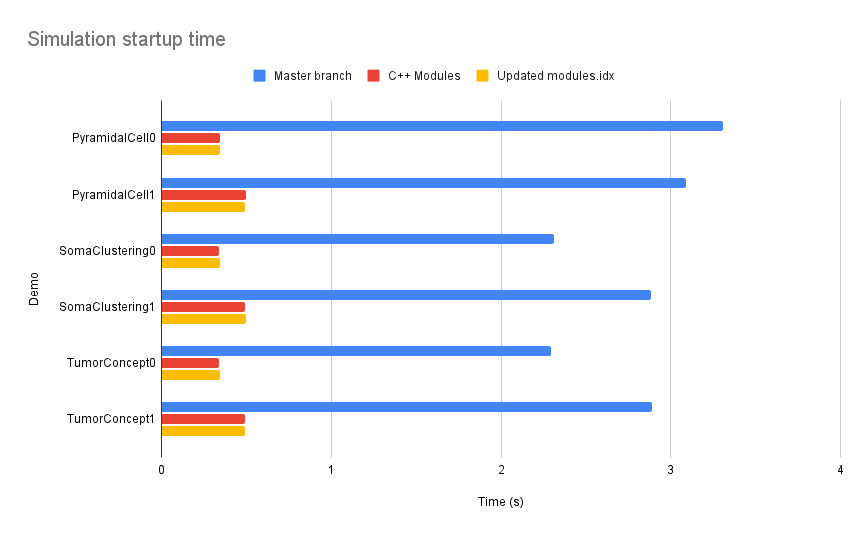
| The results have been promising, showcasing significant performance gains. Benchmarking revealed | ||
| improvements ranging from 18% reduction in peak memory usage with the default modules.idx to 25% with the | ||
| updated one. |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Add information on the number of agents in the benchmark simulations. The higher the number of agents, the lower the reduction in peak memory consumption.
Explain what models.idx is and which kind of improvements are in the updated one. In general, explain terms before you use them, such that readers that are new to your project understand.
| Cling’s parsing overhead | ||
|  | ||
|
|
||
| As expected, the simulation time did not show an appreciable improvement. However, in the |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Explain that no change in simulation runtime is expected, because the C++ modules affect only the startup phase.
| caused compatibility issues with the main build system. Also, there is a problem with the Jupyter notebooks. | ||
| Resolving these issues and finalizing the integration of C++ Modules will be essential for ensuring long-term stability and reliability. | ||
|
|
||
| Looking ahead, further optimizations are planned, including potential module-based optimizations for BioDynaMo’s |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
What exactly do you mean with module-based optimizations for BDM's core components?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
For now we are generating one big module (biodynamo.pcm). The goal is to split that into smaller ones to load them on-demand.
Project: Improving performance of BioDynaMo using ROOT C++ Modules