-
Notifications
You must be signed in to change notification settings - Fork 317
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Add Isaac Morales blog #1606
Add Isaac Morales blog #1606
Changes from all commits
File filter
Filter by extension
Conversations
Jump to
Diff view
Diff view
There are no files selected for viewing
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,135 @@ | ||
| --- | ||
| project: CERN-HSF | ||
| title: "Wrapping up GSoC'24: Improving performance of BioDynaMo using ROOT C++ Modules" | ||
| layout: blog_post | ||
| intro: "This project, part of Google Summer of Code 2024, aims to reduce the header parsing in BioDynaMo using the ROOT C++ Modules" | ||
| author: Isaac Morales Santana | ||
| photo: blog_authors/IsaacMorales.jpg | ||
| logo: CERN-HSF-GSoC-logo.png | ||
| date: 2024-10-17 | ||
| year: 2024 | ||
| tags: gsoc root cmake c++ | ||
| --- | ||
|
|
||
| ### Introduction | ||
|
|
||
| I am Isaac Morales, a Computer Engineering student at the University of Granada, Spain. | ||
| This summer I had the opportunity to participate in Google Summer of Code 2024. My project | ||
| revolved around enhancing BioDynaMo's performance using the ROOT C++ Modules. | ||
|
|
||
| **Mentors**: Vassil Vassilev, Lukas Breitwieser. | ||
|
|
||
|
|
||
| ### Project overview | ||
|
|
||
| BioDynaMo is an agent-based simulation platform designed to facilitate complex simulations, | ||
| particularly in fields like cancer research, epidemiology, and social sciences. It leverages | ||
| ROOT—a framework widely used in high-energy physics—for statistical analysis, random number | ||
| generation, C++ Jupyter notebooks, and I/O operations. However, enhancing BioDynaMo’s performance | ||
| remains a key challenge. This is where this Google Summer of Code 2024 (GSoC ‘24) project comes | ||
| into play, focusing on optimizing the platform through ROOT C++ Modules. | ||
|
|
||
| ### The Challenge: Performance Bottlenecks in BioDynaMo | ||
| BioDynaMo’s reflection system, which utilizes Cling (an interactive C++ interpreter from ROOT), | ||
| experiences significant runtime performance and memory usage issues. Specifically, parts of the system | ||
| that rely on reflection information, such as simulation initialization, are particularly slow. | ||
| At startup, the reflection subsystem has to be launched, which involves repeated parsing of library descriptors and class definitions [[1]](#1). | ||
| This process consumes substantial memory and time, especially for smaller simulations with few agents, | ||
| where more time is spent on initialization than on actual simulation steps. | ||
|
|
||
|
|
||
| ### The Solution: Integrating ROOT C++ Modules. | ||
| The primary goal of the GSoC project was to integrate ROOT’s C++ Modules into BioDynaMo to minimize | ||
| the performance overhead caused by Cling's repeated parsing of headers and descriptors. Without C++ modules, | ||
| Cling must load dictionaries and parse large libraries repeatedly at every startup. This dictionary loading process | ||
| introduces delays, as Cling resolves types and performs other initialization tasks. | ||
|
|
||
|
|
||
| #### Cling Startup and Dictionary Loading Without C++ Modules | ||
| When Cling initializes without C++ modules, it parses the header files and dictionaries of the necessary libraries from scratch. | ||
| This parsing process involves reading the contents of each header file, resolving dependencies, and loading the types | ||
| and methods defined in the libraries into Cling's reflection system. The "repeated parsing" refers to this process occurring | ||
| every time the simulation starts, since there is no cached or precompiled version of the libraries' reflection data. | ||
| Thus, each startup incurs a significant performance cost, especially for larger libraries. | ||
|
|
||
| #### How C++ Modules Solve These Challenges | ||
| C++ Modules fundamentally change how code is parsed and loaded by introducing a precompiled module format that Cling can load directly. | ||
| Instead of parsing all the header files at startup, the necessary library information is encapsulated in binary files (the modules). | ||
| When the simulation starts, Cling reads the precompiled modules, which contain reflection information, class layouts, | ||
| and other metadata required to run the simulation. This eliminates the need for repeated parsing and significantly reduces both memory consumption and startup time. | ||
| As a result, the overall performance—particularly at startup—is vastly improved. | ||
|
|
||
| By "overall performance," I refer specifically to the reduction in both memory overhead and the time required to initialize a simulation. | ||
| While the runtime of the simulation itself remains unaffected, the time it takes to get the simulation running is now much shorter, | ||
| which is critical in scenarios where multiple small simulations are run. | ||
|
|
||
| ### Promising Results | ||
|
|
||
|
|
||
| ### Benchmarking and Memory Improvements | ||
| To assess the effectiveness of the changes, we conducted benchmark simulations | ||
|
|
||
| The updated `modules.idx` file, which is the Global Module Index, plays a critical role in these optimizations. | ||
| It maintains a mapping of all available modules and ensures that Cling can efficiently load the correct modules at startup. We can add BioDynaMo modules to this index using ```root.exe -l -b -q``` after the module generation. | ||
| In the updated version, improvements include faster module lookups and better memory management during the startup phase. | ||
|
|
||
| This further contributes to the reduction in initialization time and memory overhead. | ||
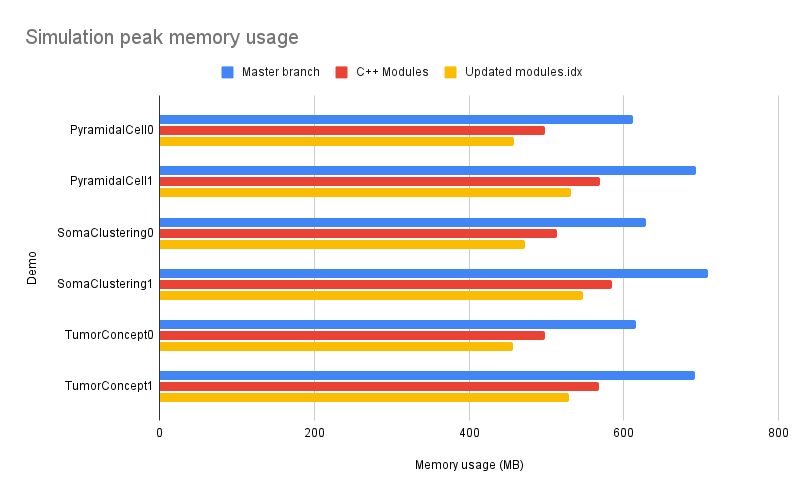
| The results have been promising, showcasing significant performance gains. Benchmarking revealed | ||
| improvements ranging from 18% reduction in peak memory usage with the default modules.idx to 25% with the | ||
| updated one. | ||
|
Comment on lines
+77
to
+79
There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. Add information on the number of agents in the benchmark simulations. The higher the number of agents, the lower the reduction in peak memory consumption. |
||
|  | ||
|
|
||
|
|
||
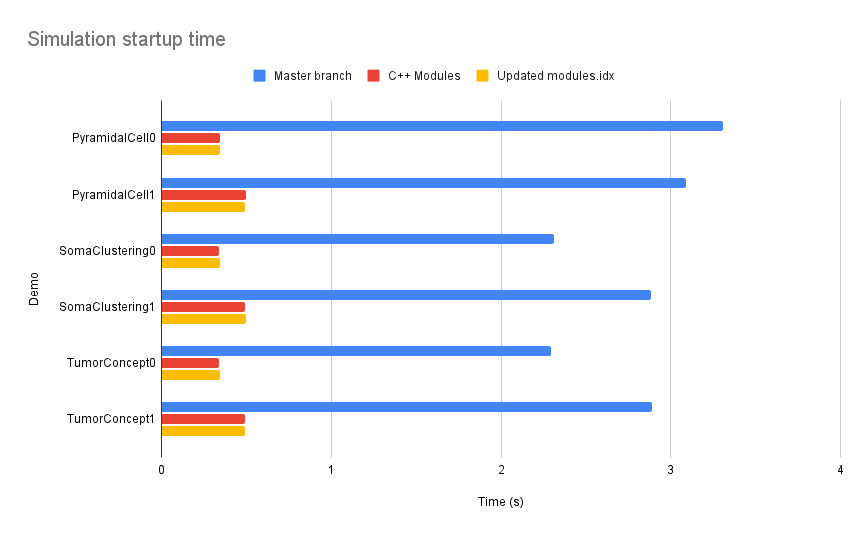
| Moreover, the startup phase saw an impressive 80% reduction in time, thanks to the optimized | ||
| handling of header parsing. That highlights the efficiency of C++ Modules in minimizing | ||
| Cling’s parsing overhead | ||
|  | ||
|
|
||
|
|
||
| ### Impact on Simulation Runtime | ||
| It's important to note that these changes specifically target the startup phase of the simulation. | ||
| No significant change in simulation runtime is expected, as the improvements are focused on reducing the overhead associated with loading and parsing libraries at startup. | ||
| Once the simulation is running, the computational tasks remain unchanged, meaning the performance benefits primarily affect the time it takes to begin the simulation, not the duration of the simulation itself. | ||
|
|
||
| ## What I did?: | ||
| 1. **Reworking CMake Rules:** The project incorporated ROOT and another packages | ||
| efficiently using FetchContent, modifying CMake rules accordingly (e.g., PR [#365](https://github.com/BioDynaMo/biodynamo/pull/365) | ||
| and [#387](https://github.com/BioDynaMo/biodynamo/pull/387), both merged) | ||
| 2. **Replacing genreflex with rootcling:** This switch was crucial to enable C++ Modules and | ||
| streamline the generation of reflection information (PR [#379](https://github.com/BioDynaMo/biodynamo/pull/379)) | ||
| 3. **C++ Modules changes** Among other things, I used automatic generation for the module map with relative paths, | ||
| modified the `selection.xml` file to support the new dictionaries and fixed headers with missing includes (PR [#385](https://github.com/BioDynaMo/biodynamo/pull/385). | ||
| 4. **Updated some CI workflows** I fixed some failing workflows in PR [#377](https://github.com/BioDynaMo/biodynamo/pull/377). | ||
| Also, I did some minor changes in some flags in PR's [#378](https://github.com/BioDynaMo/biodynamo/pull/378) and [#367](https://github.com/BioDynaMo/biodynamo/pull/367) | ||
|
|
||
|
|
||
| ### Future Steps and Challenges Ahead | ||
| Despite these advances, several challenges remain. PR's #365 is ready to merge and #385 needs some changes. For instance, memory leaks have been observed when using the new | ||
| `ROOT_GENERATE_DICTIONARY`, even with C++ Modules disabled. Additionally, the build system for individual demos has | ||
| caused compatibility issues with the main build system. Also, there is a problem with the Jupyter notebooks. | ||
| Resolving these issues and finalizing the integration of C++ Modules will be essential for ensuring long-term stability and reliability. | ||
|
|
||
| Looking ahead, further optimizations are planned, including potential module-based optimizations for BioDynaMo’s | ||
|
There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. What exactly do you mean with module-based optimizations for BDM's core components? There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. For now we are generating one big module (biodynamo.pcm). The goal is to split that into smaller ones to load them on-demand. |
||
| core components. At this time, only one big module is generated (biodynamo.pcm). Our goal is to split this module into smaller ones to load them on-demand, | ||
| improving even more the peak memory usage reduction. Collaboration with the BioDynaMo team continues, with upcoming meetings scheduled | ||
| to align efforts and resolve outstanding issues. | ||
|
|
||
| ### Conclusion | ||
| The integration of ROOT C++ Modules into BioDynaMo represents a significant step forward in improving the platform's performance, particularly in terms of startup time and memory consumption. | ||
| By addressing the root cause of inefficiencies in the reflection system, we have achieved a more streamlined and efficient simulation initialization process. | ||
| Future work could focus on further optimizations, but the foundation laid by this project promises a more scalable and efficient BioDynaMo platform. | ||
|
|
||
|
|
||
| ### Related Links | ||
|
|
||
| - [ROOT website](https://root.cern) | ||
| - [BioDynaMo website](https://www.biodynamo.org/) | ||
| - [My GitHub Profile](https://github.com/imorlxs) | ||
|
|
||
imorlxs marked this conversation as resolved.
Show resolved
Hide resolved
|
||
| ### References | ||
| - <a id="1">[1]</a> | ||
| [Calibration of stochastic, agent-based neuron growth models with approximate Bayesian computation](https://doi.org/10.1007/s00285-024-02144-2) | ||
| - [BioDynaMo: a modular platform for high-performance agent-based simulation](https://doi.org/10.1093/bioinformatics/btab649) | ||
| - [High-Performance and Scalable Agent-Based Simulation with BioDynaMo](https://doi.org/10.1145/3572848.3577480) | ||
|
|
||
|
|
||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Be more precise in explaining which parts of BioDynaMo need performance improvements and say why.
e.g.: the listed functionalities rely on reflection information; launching the reflection subsystem on simulation startup requires a significant amount of time and memory.
Cite this paper (https://link.springer.com/article/10.1007/s00285-024-02144-2), which describes the performance issue at startup.
Your work addresses this issue fundamentally at its root, while the paper above uses a workaround (yet effective) to improve performance.