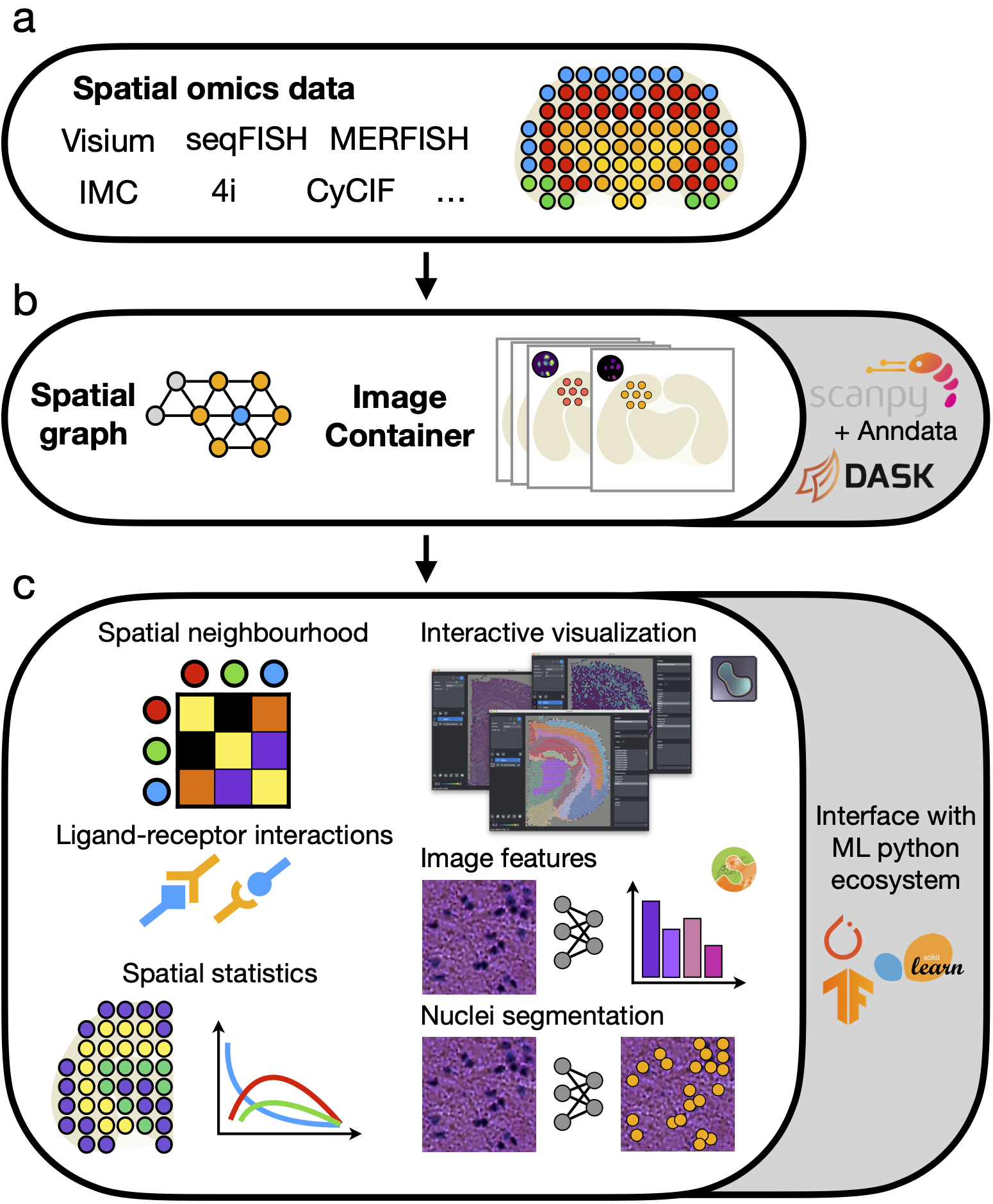
Squidpy is the scverse toolkit for scalable analysis and visualization of spatial molecular data. It builds on scanpy and anndata, providing streamlined APIs for feature extraction, spatial statistics, and interactive exploration of tissue sections together with microscopy images.
Head over to the documentation for installation instructions, tutorials, how-to guides, and reference material.
We recommend running Squidpy on a recent Linux or macOS system with Python ≥3.11, but it also works on Windows via WSL.
Install from PyPI with:
pip install squidpyor from conda-forge:
conda install -c conda-forge squidpyTo get optional dependencies required for the napari-based interactive plotting APIs, install the interactive extra:
pip install 'squidpy[interactive]'- Build and analyze spatial neighbor graphs directly from Visium, Slide-seq, Xenium, and other spatial omics assays.
- Compute spatial statistics for cell types and genes, including neighborhood enrichment, co-occurrence, and Moran's I.
- Efficiently store, featurize, and visualize high-resolution tissue microscopy images via scikit-image.
- Explore annotated datasets interactively with napari and scverse visualization tooling.
Contributions are welcome! Please read the contributing guide for instructions on setting up your environment, running tests, and submitting pull requests.
If you use Squidpy in your research, cite the original publication:
@article{palla:22,
author = {Palla, Giovanni and Spitzer, Hannah and Klein, Michal and Fischer, David and Schaar, Anna Christina
and Kuemmerle, Louis Benedikt and Rybakov, Sergei and Ibarra, Ignacio L. and Holmberg, Olle
and Virshup, Isaac and Lotfollahi, Mohammad and Richter, Sabrina and Theis, Fabian J.},
title = {Squidpy: a scalable framework for spatial omics analysis},
journal = {Nature Methods},
year = {2022},
month = {Feb},
volume = {19},
number = {2},
pages = {171--178},
issn = {1548-7105},
doi = {10.1038/s41592-021-01358-2},
}Squidpy is part of the scverse® project (website, governance) and is fiscally sponsored by NumFOCUS. Please consider making a tax-deductible donation to help the project pay for developer time, professional services, travel, workshops, and a variety of other needs.